Alexander Whatley
Improving RNA Secondary Structure Design using Deep Reinforcement Learning
Nov 05, 2021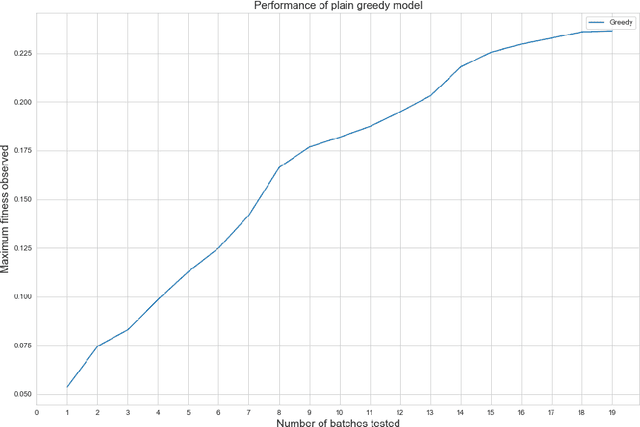
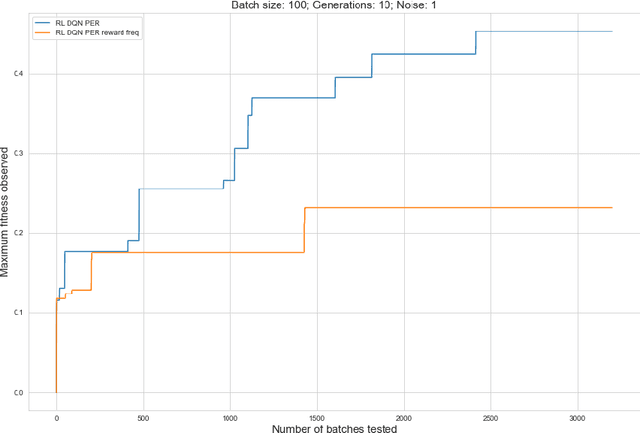
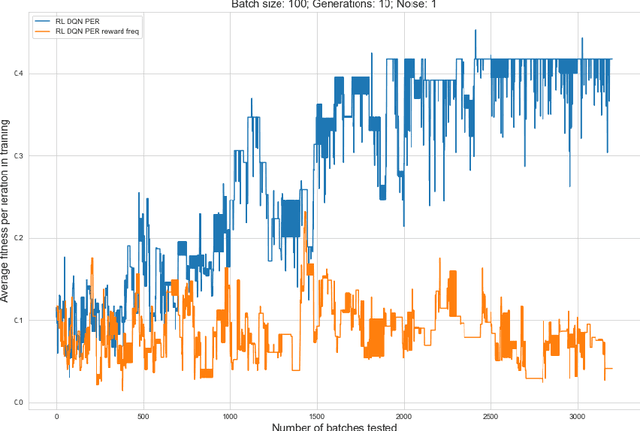
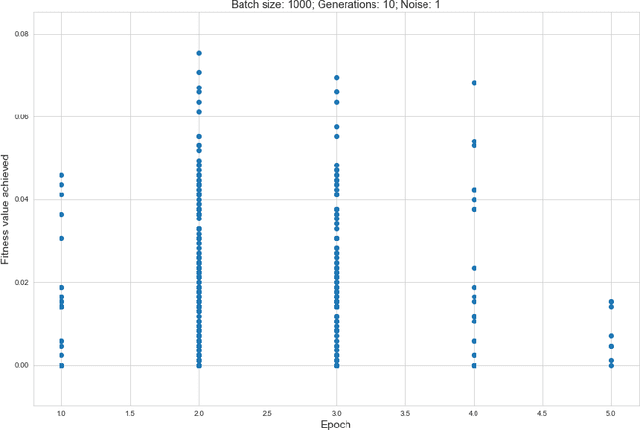
Abstract:Rising costs in recent years of developing new drugs and treatments have led to extensive research in optimization techniques in biomolecular design. Currently, the most widely used approach in biomolecular design is directed evolution, which is a greedy hill-climbing algorithm that simulates biological evolution. In this paper, we propose a new benchmark of applying reinforcement learning to RNA sequence design, in which the objective function is defined to be the free energy in the sequence's secondary structure. In addition to experimenting with the vanilla implementations of each reinforcement learning algorithm from standard libraries, we analyze variants of each algorithm in which we modify the algorithm's reward function and tune the model's hyperparameters. We show results of the ablation analysis that we do for these algorithms, as well as graphs indicating the algorithm's performance across batches and its ability to search the possible space of RNA sequences. We find that our DQN algorithm performs by far the best in this setting, contrasting with, in which PPO performs the best among all tested algorithms. Our results should be of interest to those in the biomolecular design community and should serve as a baseline for future experiments involving machine learning in molecule design.
AdaLead: A simple and robust adaptive greedy search algorithm for sequence design
Oct 05, 2020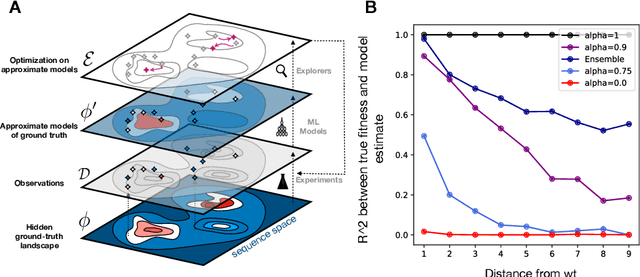
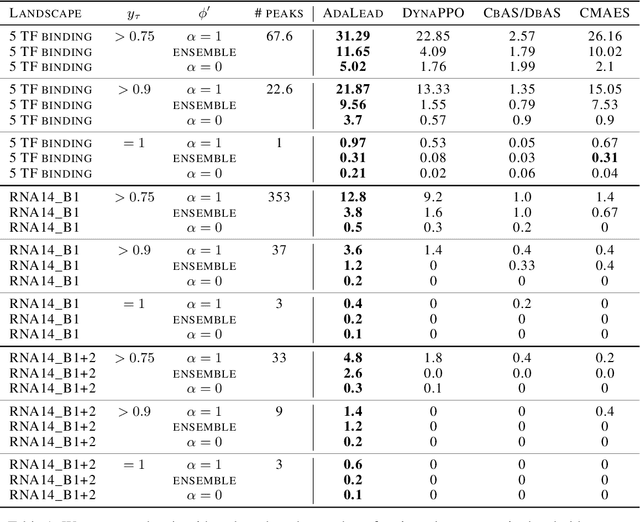
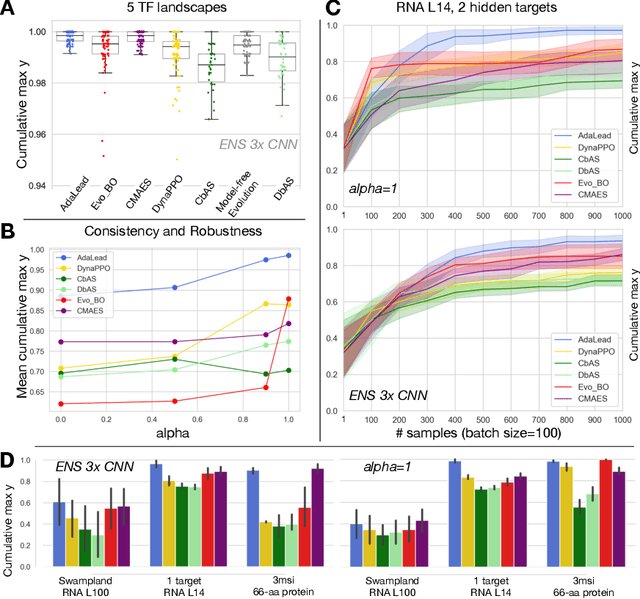
Abstract:Efficient design of biological sequences will have a great impact across many industrial and healthcare domains. However, discovering improved sequences requires solving a difficult optimization problem. Traditionally, this challenge was approached by biologists through a model-free method known as "directed evolution", the iterative process of random mutation and selection. As the ability to build models that capture the sequence-to-function map improves, such models can be used as oracles to screen sequences before running experiments. In recent years, interest in better algorithms that effectively use such oracles to outperform model-free approaches has intensified. These span from approaches based on Bayesian Optimization, to regularized generative models and adaptations of reinforcement learning. In this work, we implement an open-source Fitness Landscape EXploration Sandbox (FLEXS: github.com/samsinai/FLEXS) environment to test and evaluate these algorithms based on their optimality, consistency, and robustness. Using FLEXS, we develop an easy-to-implement, scalable, and robust evolutionary greedy algorithm (AdaLead). Despite its simplicity, we show that AdaLead is a remarkably strong benchmark that out-competes more complex state of the art approaches in a variety of biologically motivated sequence design challenges.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge