Alberto Signoroni
University of Brescia
Fighting the scanner effect in brain MRI segmentation with a progressive level-of-detail network trained on multi-site data
Nov 04, 2022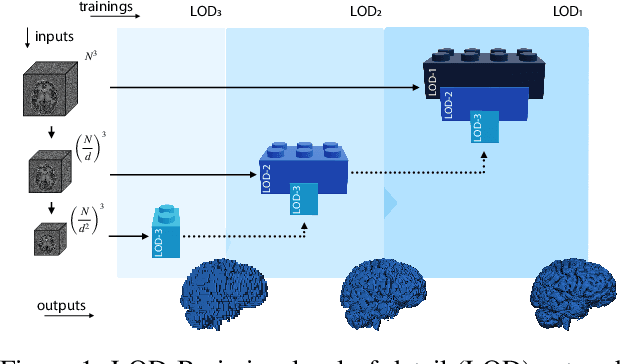

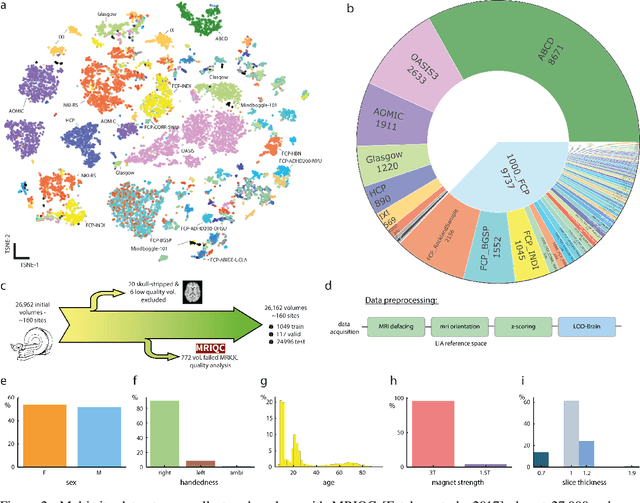
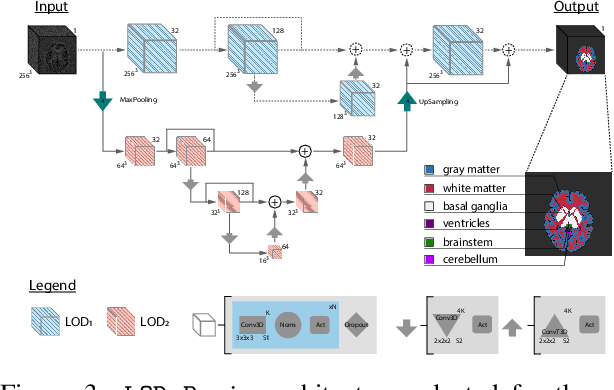
Abstract:Many clinical and research studies of the human brain require an accurate structural MRI segmentation. While traditional atlas-based methods can be applied to volumes from any acquisition site, recent deep learning algorithms ensure very high accuracy only when tested on data from the same sites exploited in training (i.e., internal data). The performance degradation experienced on external data (i.e., unseen volumes from unseen sites) is due to the inter-site variabilities in intensity distributions induced by different MR scanner models, acquisition parameters, and unique artefacts. To mitigate this site-dependency, often referred to as the scanner effect, we propose LOD-Brain, a 3D convolutional neural network with progressive levels-of-detail (LOD) able to segment brain data from any site. Coarser network levels are responsible to learn a robust anatomical prior useful for identifying brain structures and their locations, while finer levels refine the model to handle site-specific intensity distributions and anatomical variations. We ensure robustness across sites by training the model on an unprecedented rich dataset aggregating data from open repositories: almost 27,000 T1w volumes from around 160 acquisition sites, at 1.5 - 3T, from a population spanning from 8 to 90 years old. Extensive tests demonstrate that LOD-Brain produces state-of-the-art results, with no significant difference in performance between internal and external sites, and robust to challenging anatomical variations. Its portability opens the way for large scale application across different healthcare institutions, patient populations, and imaging technology manufacturers. Code, model, and demo are available at the project website.
End-to-end learning for semiquantitative rating of COVID-19 severity on Chest X-rays
Jun 08, 2020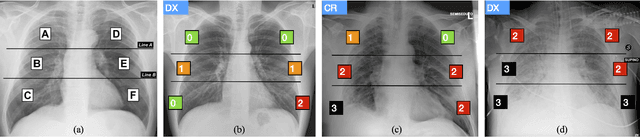
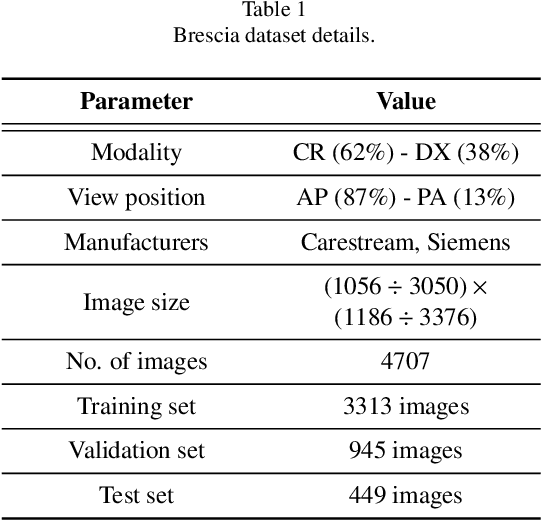
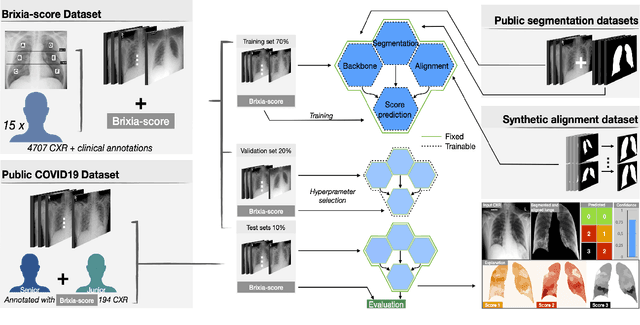
Abstract:In this work we designed an end-to-end deep learning architecture for predicting, on Chest X-rays images (CRX), a multi-regional score conveying the degree of lung compromise in COVID-19 patients. Such semiquantitative scoring system, namely Brixia-score, was applied in serial monitoring of such patients, showing significant prognostic value, in one of the hospitals that experienced one of the highest pandemic peaks in Italy. To solve such a challenging visual task, we adopt a weakly supervised learning strategy structured to handle different tasks (segmentation, spatial alignment, and score estimation) trained with a "from part to whole" procedure involving different datasets. In particular, we exploited a clinical dataset of almost 5,000 CXR annotated images collected in the same hospital. Our BS-Net demonstrated self-attentive behavior and a high degree of accuracy in all processing stages. Through inter-rater agreement tests and a gold standard comparison, we were able to show that our solution outperforms single human annotators in rating accuracy and consistency, thus supporting the possibility of using this tool in contexts of computer-assisted monitoring. Highly resolved (super-pixel level) explainability maps were also generated, with an original technique, to visually help the understanding of the network activity on the lung areas. We eventually tested the performance robustness of our model on a variegated public COVID-19 dataset, for which we also provide Brixia-score annotations, observing good direct generalization and fine-tuning capabilities that favorably highlight the portability of BS-Net in other clinical settings.
CEREBRUM: a fast and fully-volumetric Convolutional Encoder-decodeR for weakly-supervised sEgmentation of BRain strUctures from out-of-the-scanner MRI
Sep 26, 2019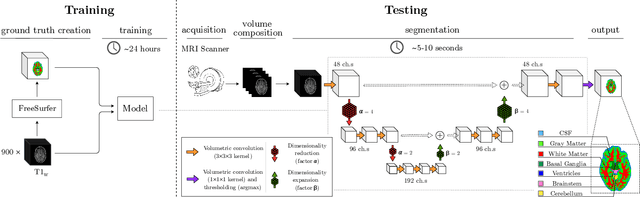
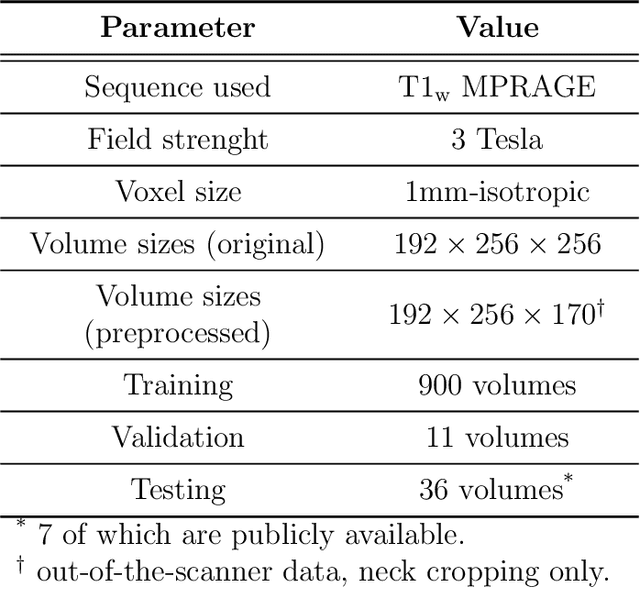
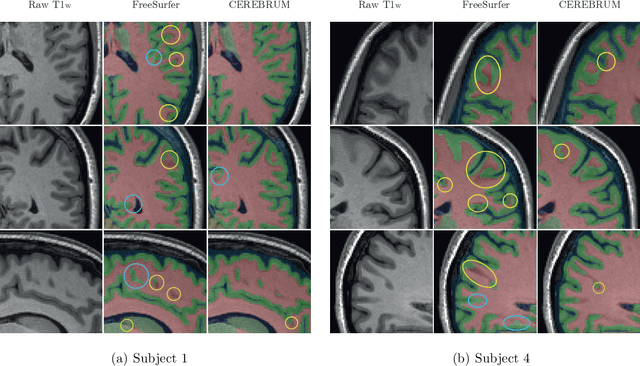
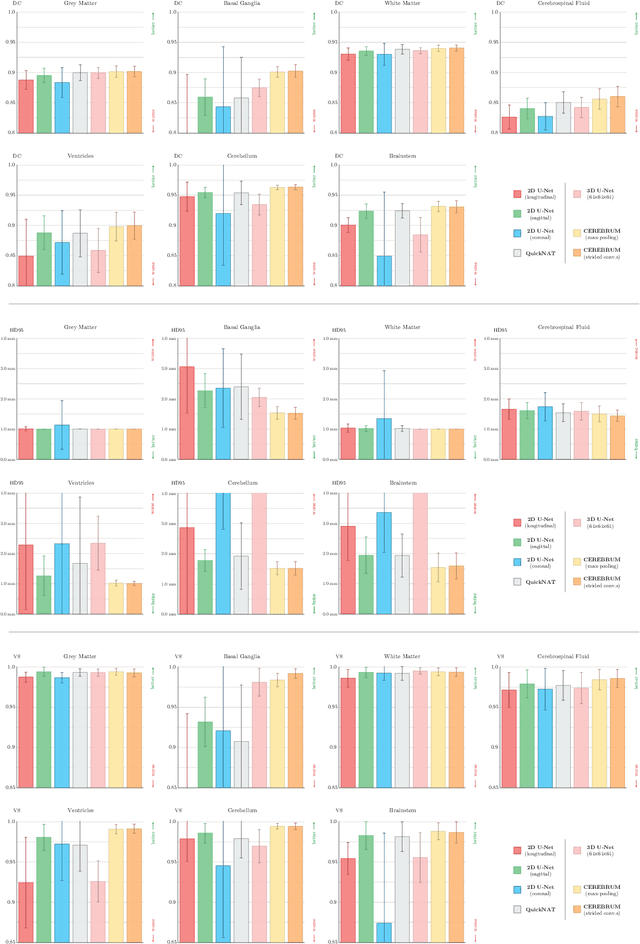
Abstract:Many functional and structural neuroimaging studies call for accurate morphometric segmentation of different brain structures starting from image intensity values of MRI scans. Current automatic (multi-) atlas-based segmentation strategies often lack accuracy on difficult-to-segment brain structures and, since these methods rely on atlas-to-scan alignment, they may take long processing times. Recently, methods deploying solutions based on Convolutional Neural Networks (CNNs) are making the direct analysis of out-of-the-scanner data feasible. However, current CNN-based solutions partition the test volume into 2D or 3D patches, which are processed independently. This entails a loss of global contextual information thereby negatively impacting the segmentation accuracy. In this work, we design and test an optimised end-to-end CNN architecture that makes the exploitation of global spatial information computationally tractable, allowing to process a whole MRI volume at once. We adopt a weakly supervised learning strategy by exploiting a large dataset composed by 947 out-of-the-scanner (3 Tesla T1-weighted 1mm isotropic MP-RAGE 3D sequences) MR Images. The resulting model is able to produce accurate multi-structure segmentation results in only few seconds. Different quantitative measures demonstrate an improved accuracy of our solution when compared to state-of-the-art techniques. Moreover, through a randomised survey involving expert neuroscientists, we show that subjective judgements clearly prefer our solution with respect to the widely adopted atlas-based FreeSurfer software.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge