Ahmed Fakhry
ODKE+: Ontology-Guided Open-Domain Knowledge Extraction with LLMs
Sep 04, 2025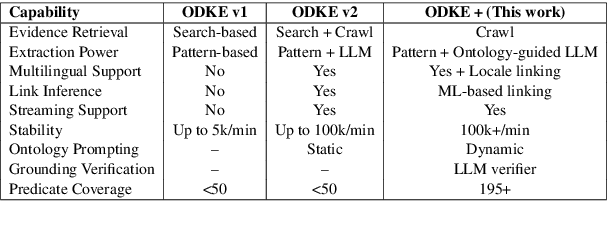
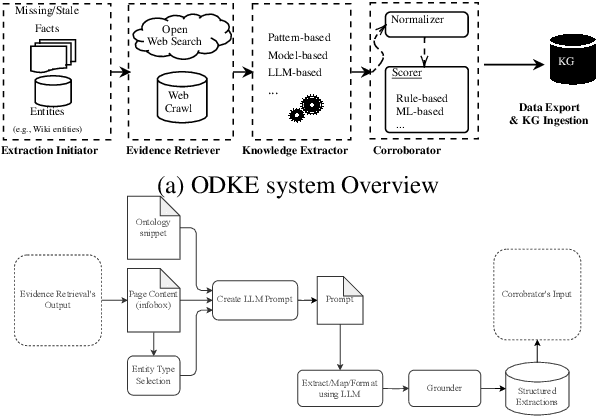
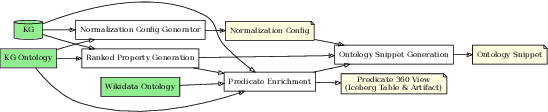

Abstract:Knowledge graphs (KGs) are foundational to many AI applications, but maintaining their freshness and completeness remains costly. We present ODKE+, a production-grade system that automatically extracts and ingests millions of open-domain facts from web sources with high precision. ODKE+ combines modular components into a scalable pipeline: (1) the Extraction Initiator detects missing or stale facts, (2) the Evidence Retriever collects supporting documents, (3) hybrid Knowledge Extractors apply both pattern-based rules and ontology-guided prompting for large language models (LLMs), (4) a lightweight Grounder validates extracted facts using a second LLM, and (5) the Corroborator ranks and normalizes candidate facts for ingestion. ODKE+ dynamically generates ontology snippets tailored to each entity type to align extractions with schema constraints, enabling scalable, type-consistent fact extraction across 195 predicates. The system supports batch and streaming modes, processing over 9 million Wikipedia pages and ingesting 19 million high-confidence facts with 98.8% precision. ODKE+ significantly improves coverage over traditional methods, achieving up to 48% overlap with third-party KGs and reducing update lag by 50 days on average. Our deployment demonstrates that LLM-based extraction, grounded in ontological structure and verification workflows, can deliver trustworthiness, production-scale knowledge ingestion with broad real-world applicability. A recording of the system demonstration is included with the submission and is also available at https://youtu.be/UcnE3_GsTWs.
GENER: A Parallel Layer Deep Learning Network To Detect Gene-Gene Interactions From Gene Expression Data
Oct 06, 2023Abstract:Detecting and discovering new gene interactions based on known gene expressions and gene interaction data presents a significant challenge. Various statistical and deep learning methods have attempted to tackle this challenge by leveraging the topological structure of gene interactions and gene expression patterns to predict novel gene interactions. In contrast, some approaches have focused exclusively on utilizing gene expression profiles. In this context, we introduce GENER, a parallel-layer deep learning network designed exclusively for the identification of gene-gene relationships using gene expression data. We conducted two training experiments and compared the performance of our network with that of existing statistical and deep learning approaches. Notably, our model achieved an average AUROC score of 0.834 on the combined BioGRID&DREAM5 dataset, outperforming competing methods in predicting gene-gene interactions.
Virufy: A Multi-Branch Deep Learning Network for Automated Detection of COVID-19
Mar 16, 2021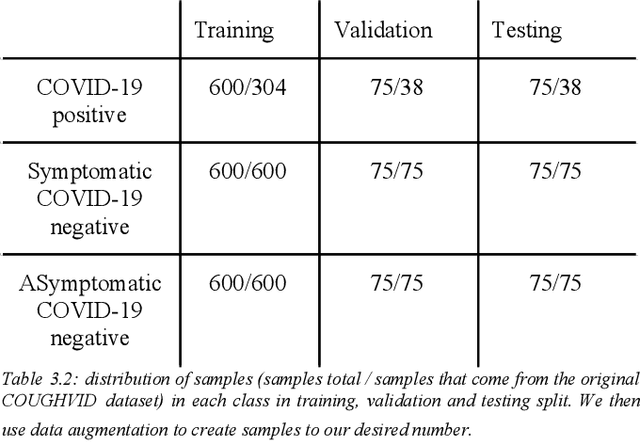
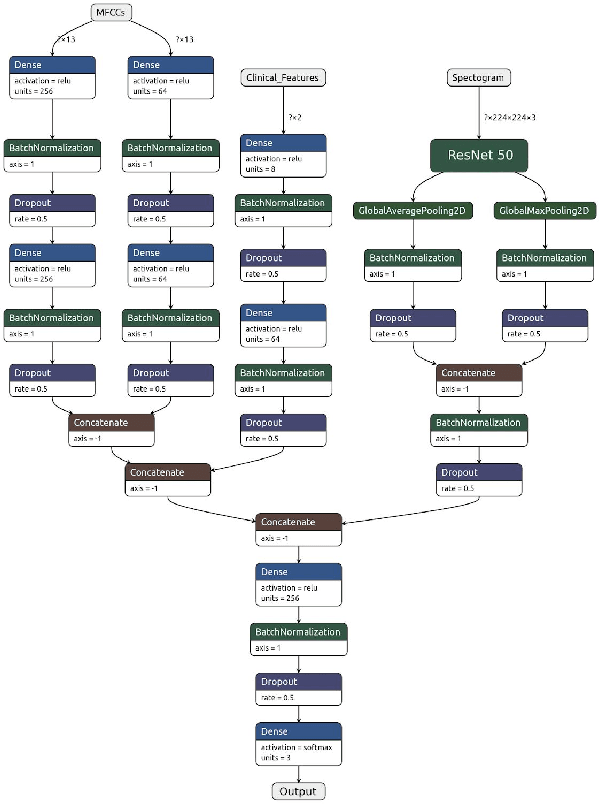
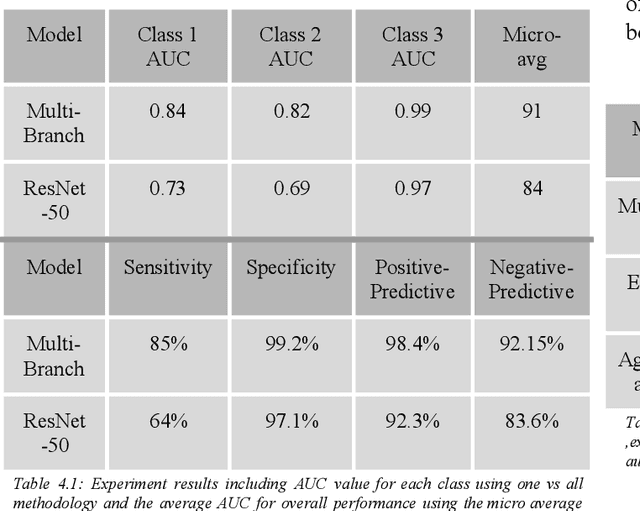
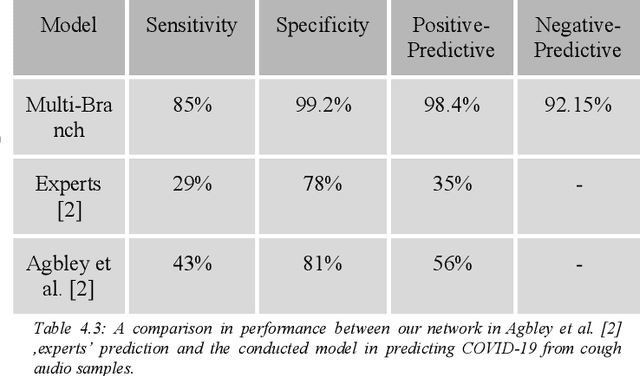
Abstract:Fast and affordable solutions for COVID-19 testing are necessary to contain the spread of the global pandemic and help relieve the burden on medical facilities. Currently, limited testing locations and expensive equipment pose difficulties for individuals trying to be tested, especially in low-resource settings. Researchers have successfully presented models for detecting COVID-19 infection status using audio samples recorded in clinical settings [5, 15], suggesting that audio-based Artificial Intelligence models can be used to identify COVID-19. Such models have the potential to be deployed on smartphones for fast, widespread, and low-resource testing. However, while previous studies have trained models on cleaned audio samples collected mainly from clinical settings, audio samples collected from average smartphones may yield suboptimal quality data that is different from the clean data that models were trained on. This discrepancy may add a bias that affects COVID-19 status predictions. To tackle this issue, we propose a multi-branch deep learning network that is trained and tested on crowdsourced data where most of the data has not been manually processed and cleaned. Furthermore, the model achieves state-of-art results for the COUGHVID dataset [16]. After breaking down results for each category, we have shown an AUC of 0.99 for audio samples with COVID-19 positive labels.
Virufy: Global Applicability of Crowdsourced and Clinical Datasets for AI Detection of COVID-19 from Cough
Dec 25, 2020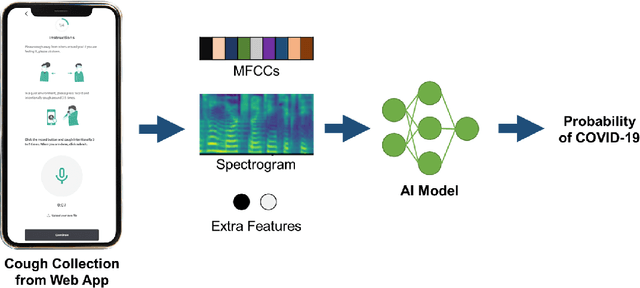
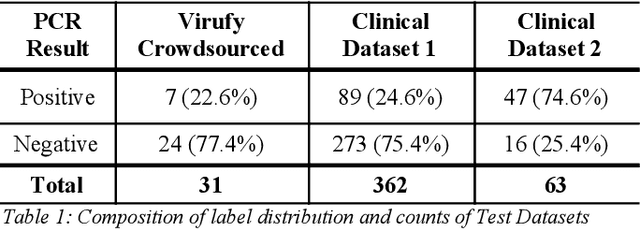
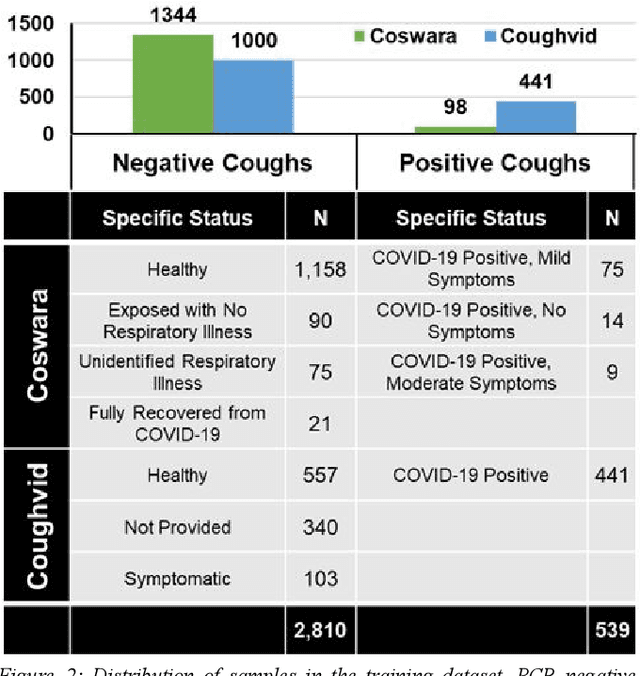
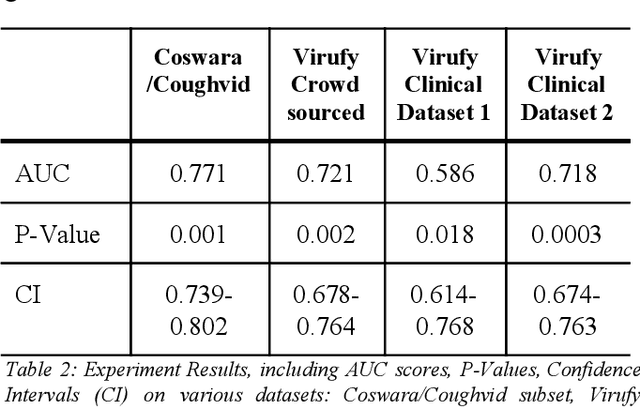
Abstract:Rapid and affordable methods of testing for COVID-19 infections are essential to reduce infection rates and prevent medical facilities from becoming overwhelmed. Current approaches of detecting COVID-19 require in-person testing with expensive kits that are not always easily accessible. This study demonstrates that crowdsourced cough audio samples recorded and acquired on smartphones from around the world can be used to develop an AI-based method that accurately predicts COVID-19 infection with an ROC-AUC of 77.1% (75.2%-78.3%). Furthermore, we show that our method is able to generalize to crowdsourced audio samples from Latin America and clinical samples from South Asia, without further training using the specific samples from those regions. As more crowdsourced data is collected, further development can be implemented using various respiratory audio samples to create a cough analysis-based machine learning (ML) solution for COVID-19 detection that can likely generalize globally to all demographic groups in both clinical and non-clinical settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge