Abhishek Mall
Observation of Aerosolization-induced Morphological Changes in Viral Capsids
Jul 16, 2024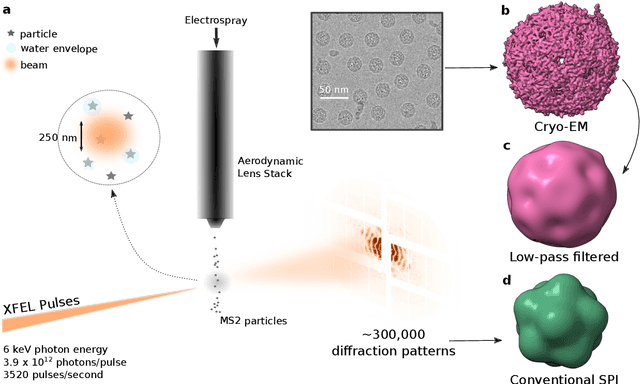
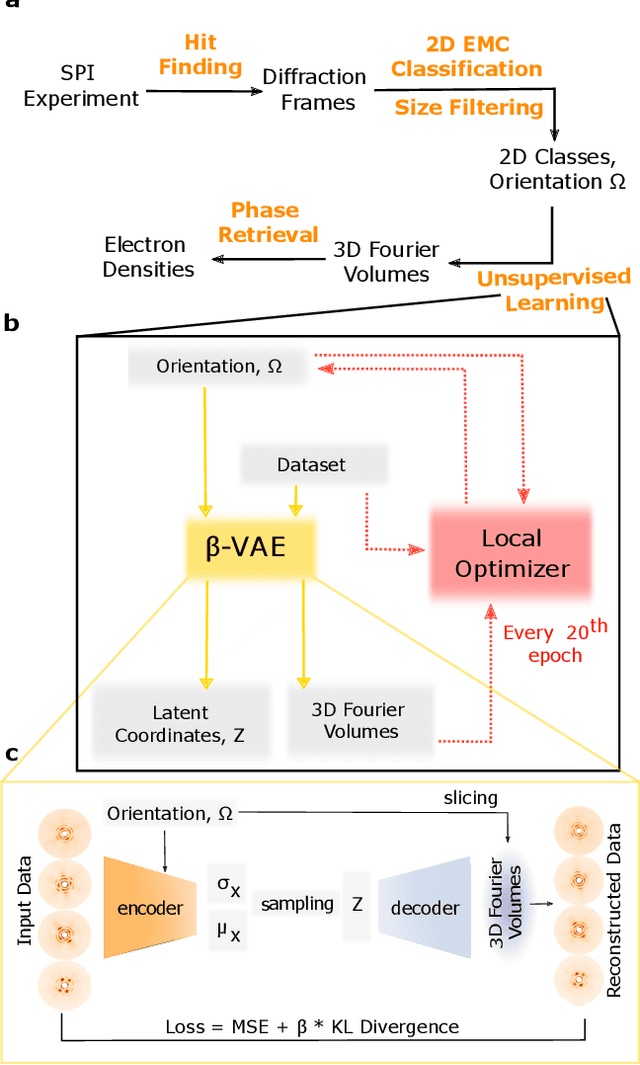
Abstract:Single-stranded RNA viruses co-assemble their capsid with the genome and variations in capsid structures can have significant functional relevance. In particular, viruses need to respond to a dehydrating environment to prevent genomic degradation and remain active upon rehydration. Theoretical work has predicted low-energy buckling transitions in icosahedral capsids which could protect the virus from further dehydration. However, there has been no direct experimental evidence, nor molecular mechanism, for such behaviour. Here we observe this transition using X-ray single particle imaging of MS2 bacteriophages after aerosolization. Using a combination of machine learning tools, we classify hundreds of thousands of single particle diffraction patterns to learn the structural landscape of the capsid morphology as a function of time spent in the aerosol phase. We found a previously unreported compact conformation as well as intermediate structures which suggest an incoherent buckling transition which does not preserve icosahedral symmetry. Finally, we propose a mechanism of this buckling, where a single 19-residue loop is destabilised, leading to the large observed morphology change. Our results provide experimental evidence for a mechanism by which viral capsids protect themselves from dehydration. In the process, these findings also demonstrate the power of single particle X-ray imaging and machine learning methods in studying biomolecular structural dynamics.
Deep learning-based variational autoencoder for classification of quantum and classical states of light
May 08, 2024Abstract:Advancements in optical quantum technologies have been enabled by the generation, manipulation, and characterization of light, with identification based on its photon statistics. However, characterizing light and its sources through single photon measurements often requires efficient detectors and longer measurement times to obtain high-quality photon statistics. Here we introduce a deep learning-based variational autoencoder (VAE) method for classifying single photon added coherent state (SPACS), single photon added thermal state (SPACS), mixed states between coherent/SPACS and thermal/SPATS of light. Our semisupervised learning-based VAE efficiently maps the photon statistics features of light to a lower dimension, enabling quasi-instantaneous classification with low average photon counts. The proposed VAE method is robust and maintains classification accuracy in the presence of losses inherent in an experiment, such as finite collection efficiency, non-unity quantum efficiency, finite number of detectors, etc. Additionally, leveraging the transfer learning capabilities of VAE enables successful classification of data of any quality using a single trained model. We envision that such a deep learning methodology will enable better classification of quantum light and light sources even in the presence of poor detection quality.
Holographic single particle imaging for weakly scattering, heterogeneous nanoscale objects
Oct 19, 2022
Abstract:Single particle imaging (SPI) at X-ray free electron lasers (XFELs) is a technique to determine the 3D structure of nanoscale objects like biomolecules from a large number of diffraction patterns of copies of these objects in random orientations. Millions of low signal-to-noise diffraction patterns with unknown orientation are collected during an X-ray SPI experiment. The patterns are then analyzed and merged using a reconstruction algorithm to retrieve the full 3D-structure of particle. The resolution of reconstruction is limited by background noise, signal-to-noise ratio in diffraction patterns and total amount of data collected. We recently introduced a reference-enhanced holographic single particle imaging methodology [Optica 7,593-601(2020)] to collect high enough signal-to-noise and background tolerant patterns and a reconstruction algorithm to recover missing parameters beyond orientation and then directly retrieve the full Fourier model of the sample of interest. Here we describe a phase retrieval algorithm based on maximum likelihood estimation using pattern search dubbed as MaxLP, with better scalability for fine sampling of latent parameters and much better performance in the low signal limit. Furthermore, we show that structural variations within the target particle are averaged in real space, significantly improving robustness to conformational heterogeneity in comparison to conventional SPI. With these computational improvements, we believe reference-enhanced SPI is capable of reaching sub-nm resolution biomolecule imaging.
Unsupervised learning approaches to characterize heterogeneous samples using X-ray single particle imaging
Sep 13, 2021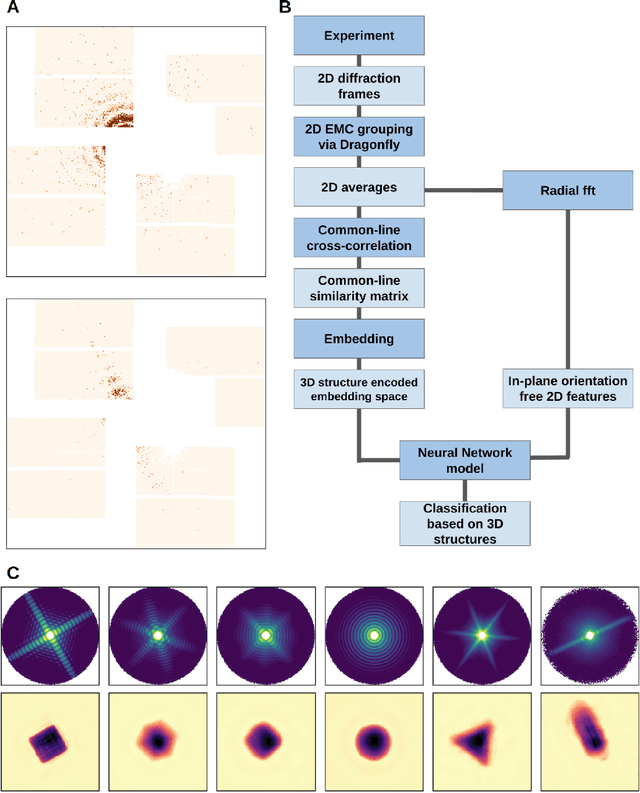
Abstract:One of the outstanding analytical problems in X-ray single particle imaging (SPI) is the classification of structural heterogeneity, which is especially difficult given the low signal-to-noise ratios of individual patterns and that even identical objects can yield patterns that vary greatly when orientation is taken into consideration. We propose two methods which explicitly account for this orientation-induced variation and can robustly determine the structural landscape of a sample ensemble. The first, termed common-line principal component analysis (PCA) provides a rough classification which is essentially parameter-free and can be run automatically on any SPI dataset. The second method, utilizing variation auto-encoders (VAEs) can generate 3D structures of the objects at any point in the structural landscape. We implement both these methods in combination with the noise-tolerant expand-maximize-compress (EMC) algorithm and demonstrate its utility by applying it to an experimental dataset from gold nanoparticles with only a few thousand photons per pattern and recover both discrete structural classes as well as continuous deformations. These developments diverge from previous approaches of extracting reproducible subsets of patterns from a dataset and open up the possibility to move beyond studying homogeneous sample sets and study open questions on topics such as nanocrystal growth and dynamics as well as phase transitions which have not been externally triggered.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge