U-Net Fixed-Point Quantization for Medical Image Segmentation
Paper and Code
Sep 09, 2019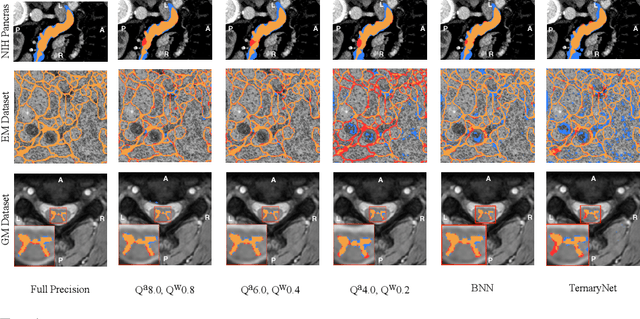
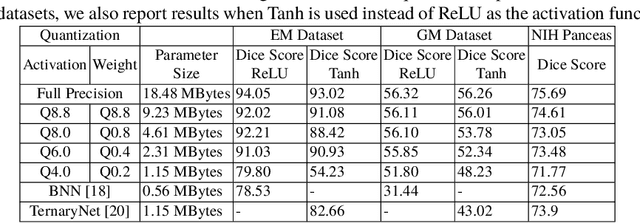
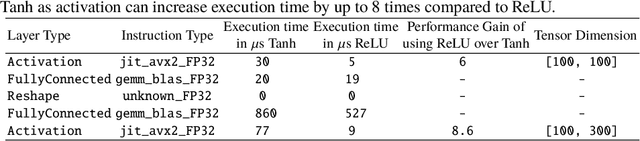
Model quantization is leveraged to reduce the memory consumption and the computation time of deep neural networks. This is achieved by representing weights and activations with a lower bit resolution when compared to their high precision floating point counterparts. The suitable level of quantization is directly related to the model performance. Lowering the quantization precision (e.g. 2 bits), reduces the amount of memory required to store model parameters and the amount of logic required to implement computational blocks, which contributes to reducing the power consumption of the entire system. These benefits typically come at the cost of reduced accuracy. The main challenge is to quantize a network as much as possible, while maintaining the performance accuracy. In this work, we present a quantization method for the U-Net architecture, a popular model in medical image segmentation. We then apply our quantization algorithm to three datasets: (1) the Spinal Cord Gray Matter Segmentation (GM), (2) the ISBI challenge for segmentation of neuronal structures in Electron Microscopic (EM), and (3) the public National Institute of Health (NIH) dataset for pancreas segmentation in abdominal CT scans. The reported results demonstrate that with only 4 bits for weights and 6 bits for activations, we obtain 8 fold reduction in memory requirements while loosing only 2.21%, 0.57% and 2.09% dice overlap score for EM, GM and NIH datasets respectively. Our fixed point quantization provides a flexible trade off between accuracy and memory requirement which is not provided by previous quantization methods for U-Net such as TernaryNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge