Transfer Learning for Clinical Time Series Analysis using Recurrent Neural Networks
Paper and Code
Jul 04, 2018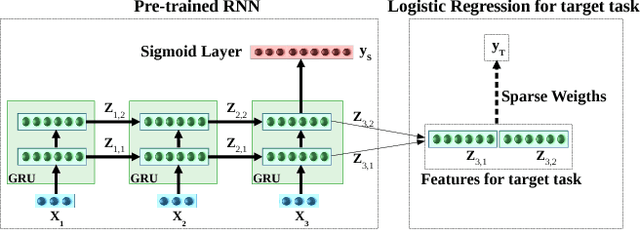

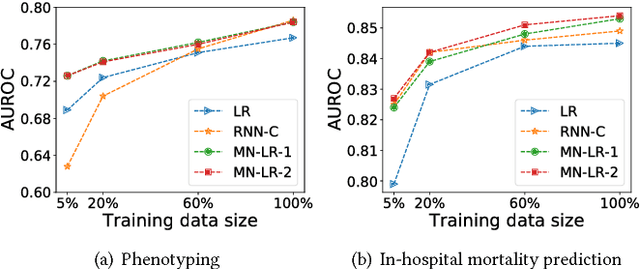

Deep neural networks have shown promising results for various clinical prediction tasks such as diagnosis, mortality prediction, predicting duration of stay in hospital, etc. However, training deep networks -- such as those based on Recurrent Neural Networks (RNNs) -- requires large labeled data, high computational resources, and significant hyperparameter tuning effort. In this work, we investigate as to what extent can transfer learning address these issues when using deep RNNs to model multivariate clinical time series. We consider transferring the knowledge captured in an RNN trained on several source tasks simultaneously using a large labeled dataset to build the model for a target task with limited labeled data. An RNN pre-trained on several tasks provides generic features, which are then used to build simpler linear models for new target tasks without training task-specific RNNs. For evaluation, we train a deep RNN to identify several patient phenotypes on time series from MIMIC-III database, and then use the features extracted using that RNN to build classifiers for identifying previously unseen phenotypes, and also for a seemingly unrelated task of in-hospital mortality. We demonstrate that (i) models trained on features extracted using pre-trained RNN outperform or, in the worst case, perform as well as task-specific RNNs; (ii) the models using features from pre-trained models are more robust to the size of labeled data than task-specific RNNs; and (iii) features extracted using pre-trained RNN are generic enough and perform better than typical statistical hand-crafted features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge