SurvMamba: State Space Model with Multi-grained Multi-modal Interaction for Survival Prediction
Paper and Code
Apr 11, 2024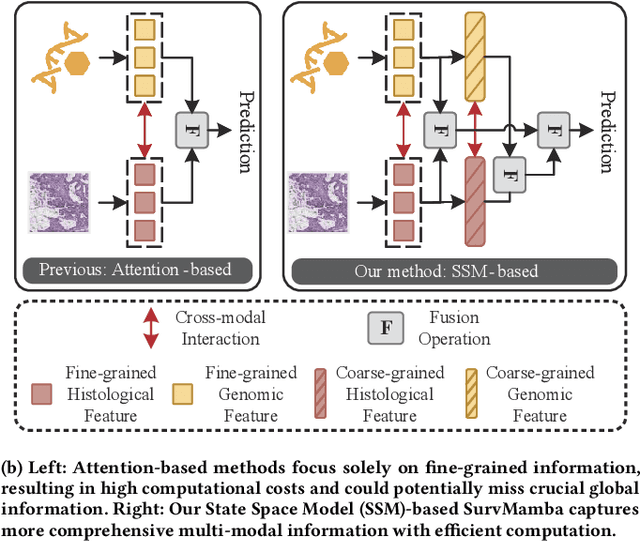
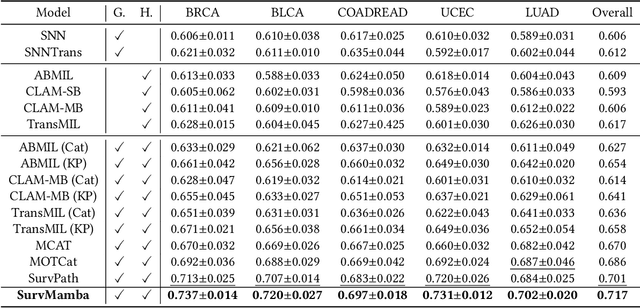
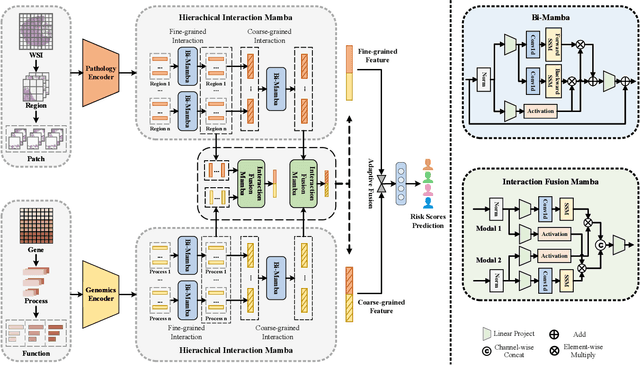
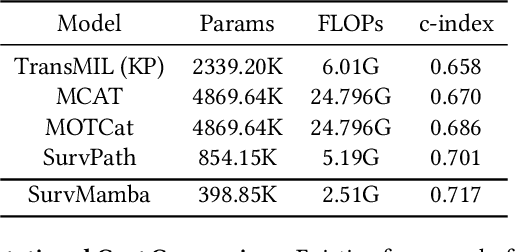
Multi-modal learning that combines pathological images with genomic data has significantly enhanced the accuracy of survival prediction. Nevertheless, existing methods have not fully utilized the inherent hierarchical structure within both whole slide images (WSIs) and transcriptomic data, from which better intra-modal representations and inter-modal integration could be derived. Moreover, many existing studies attempt to improve multi-modal representations through attention mechanisms, which inevitably lead to high complexity when processing high-dimensional WSIs and transcriptomic data. Recently, a structured state space model named Mamba emerged as a promising approach for its superior performance in modeling long sequences with low complexity. In this study, we propose Mamba with multi-grained multi-modal interaction (SurvMamba) for survival prediction. SurvMamba is implemented with a Hierarchical Interaction Mamba (HIM) module that facilitates efficient intra-modal interactions at different granularities, thereby capturing more detailed local features as well as rich global representations. In addition, an Interaction Fusion Mamba (IFM) module is used for cascaded inter-modal interactive fusion, yielding more comprehensive features for survival prediction. Comprehensive evaluations on five TCGA datasets demonstrate that SurvMamba outperforms other existing methods in terms of performance and computational cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge