Simulation-supervised deep learning for analysing organelles states and behaviour in living cells
Paper and Code
Aug 26, 2020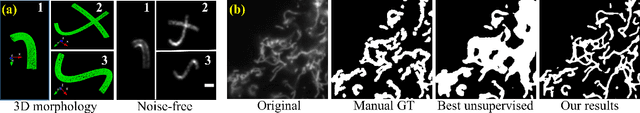
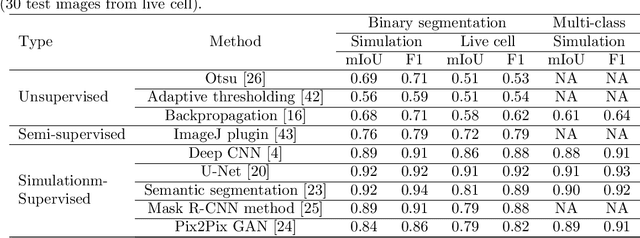

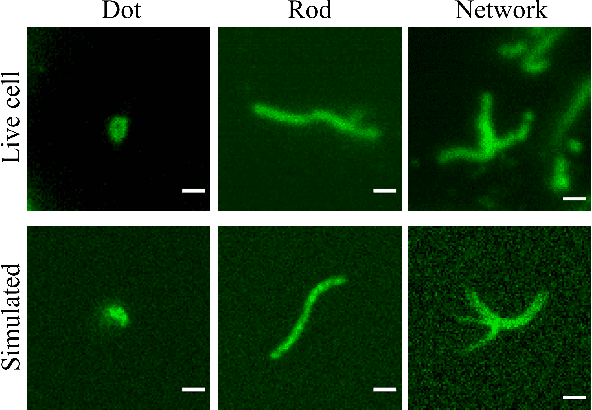
In many real-world scientific problems, generating ground truth (GT) for supervised learning is almost impossible. The causes include limitations imposed by scientific instrument, physical phenomenon itself, or the complexity of modeling. Performing artificial intelligence (AI) tasks such as segmentation, tracking, and analytics of small sub-cellular structures such as mitochondria in microscopy videos of living cells is a prime example. The 3D blurring function of microscope, digital resolution from pixel size, optical resolution due to the character of light, noise characteristics, and complex 3D deformable shapes of mitochondria, all contribute to making this problem GT hard. Manual segmentation of 100s of mitochondria across 1000s of frames and then across many such videos is not only herculean but also physically inaccurate because of the instrument and phenomena imposed limitations. Unsupervised learning produces less than optimal results and accuracy is important if inferences relevant to therapy are to be derived. In order to solve this unsurmountable problem, we bring modeling and deep learning to a nexus. We show that accurate physics based modeling of microscopy data including all its limitations can be the solution for generating simulated training datasets for supervised learning. We show here that our simulation-supervised segmentation approach is a great enabler for studying mitochondrial states and behaviour in heart muscle cells, where mitochondria have a significant role to play in the health of the cells. We report unprecedented mean IoU score of 91% for binary segmentation (19% better than the best performing unsupervised approach) of mitochondria in actual microscopy videos of living cells. We further demonstrate the possibility of performing multi-class classification, tracking, and morphology associated analytics at the scale of individual mitochondrion.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge