SELFIES: a robust representation of semantically constrained graphs with an example application in chemistry
Paper and Code
May 31, 2019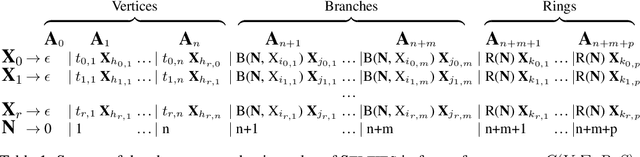
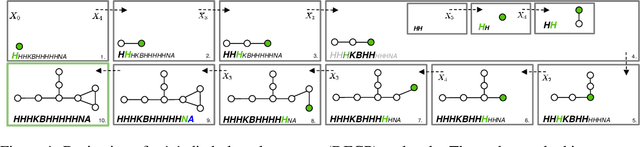
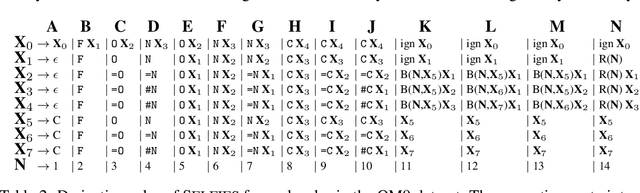
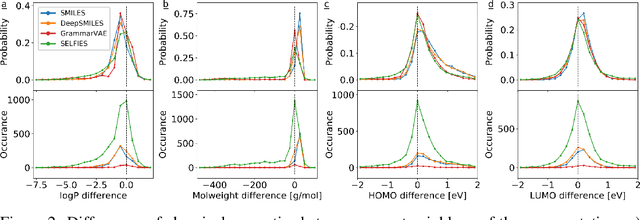
Graphs are ideal representations of complex, relational information. Their applications span diverse areas of science and engineering, such as Feynman diagrams in fundamental physics, the structures of molecules in chemistry or transport systems in urban planning. Recently, many of these examples turned into the spotlight as applications of machine learning (ML). There, common challenges to the successful deployment of ML are domain-specific constraints, which lead to semantically constrained graphs. While much progress has been achieved in the generation of valid graphs for domain- and model-specific applications, a general approach has not been demonstrated yet. Here, we present a general-purpose, sequence-based, robust representation of semantically constrained graphs, which we call SELFIES (SELF-referencIng Embedded Strings). SELFIES are based on a Chomsky type-2 grammar, augmented with two self-referencing functions. We demonstrate their applicability to represent chemical compound structures and compare them to perhaps the most popular 2D representation, SMILES, and other important baselines. We find stronger robustness against character mutations while still maintaining similar chemical properties. Even entirely random SELFIES produce semantically valid graphs in most of the cases. As feature representation in variational autoencoders, SELFIES provide a substantial improvement in the task of in reconstruction, validity, and diversity. We anticipate that SELFIES allow for direct applications in ML, without the need for domain-specific adaptation of model architectures. SELFIES are not limited to the structures of small molecules, and we show how to apply them to two other examples from the sciences: representations of DNA and interaction graphs for quantum mechanical experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge