Priberam at MESINESP Multi-label Classification of Medical Texts Task
Paper and Code
May 12, 2021
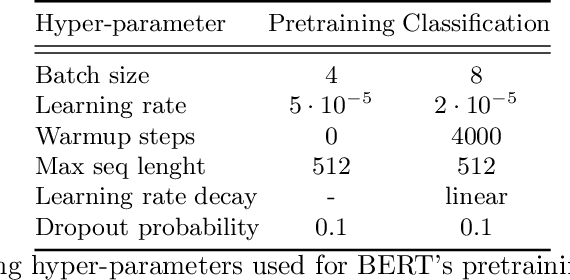

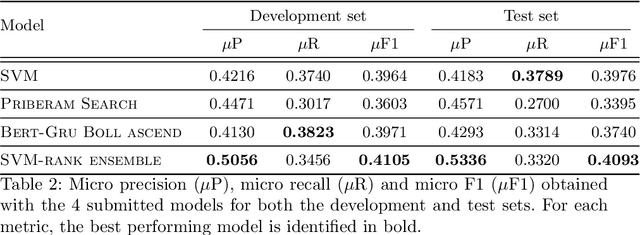
Medical articles provide current state of the art treatments and diagnostics to many medical practitioners and professionals. Existing public databases such as MEDLINE contain over 27 million articles, making it difficult to extract relevant content without the use of efficient search engines. Information retrieval tools are crucial in order to navigate and provide meaningful recommendations for articles and treatments. Classifying these articles into broader medical topics can improve the retrieval of related articles. The set of medical labels considered for the MESINESP task is on the order of several thousands of labels (DeCS codes), which falls under the extreme multi-label classification problem. The heterogeneous and highly hierarchical structure of medical topics makes the task of manually classifying articles extremely laborious and costly. It is, therefore, crucial to automate the process of classification. Typical machine learning algorithms become computationally demanding with such a large number of labels and achieving better recall on such datasets becomes an unsolved problem. This work presents Priberam's participation at the BioASQ task Mesinesp. We address the large multi-label classification problem through the use of four different models: a Support Vector Machine (SVM), a customised search engine (Priberam Search), a BERT based classifier, and a SVM-rank ensemble of all the previous models. Results demonstrate that all three individual models perform well and the best performance is achieved by their ensemble, granting Priberam the 6th place in the present challenge and making it the 2nd best team.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge