PeTailor: Improving Large Language Model by Tailored Chunk Scorer in Biomedical Triple Extraction
Paper and Code
Oct 27, 2023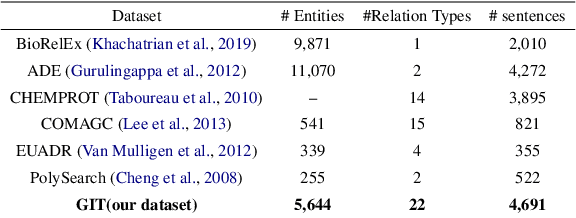



The automatic extraction of biomedical entities and their interaction from unstructured data remains a challenging task due to the limited availability of expert-labeled standard datasets. In this paper, we introduce PETAI-LOR, a retrieval-based language framework that is augmented by tailored chunk scorer. Unlike previous retrieval-augmented language models (LM) that retrieve relevant documents by calculating the similarity between the input sentence and the candidate document set, PETAILOR segments the sentence into chunks and retrieves the relevant chunk from our pre-computed chunk-based relational key-value memory. Moreover, in order to comprehend the specific requirements of the LM, PETAI-LOR adapt the tailored chunk scorer to the LM. We also introduce GM-CIHT, an expert annotated biomedical triple extraction dataset with more relation types. This dataset is centered on the non-drug treatment and general biomedical domain. Additionally, we investigate the efficacy of triple extraction models trained on general domains when applied to the biomedical domain. Our experiments reveal that PETAI-LOR achieves state-of-the-art performance on GM-CIHT
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge