Multi-task Joint Strategies of Self-supervised Representation Learning on Biomedical Networks for Drug Discovery
Paper and Code
Jan 12, 2022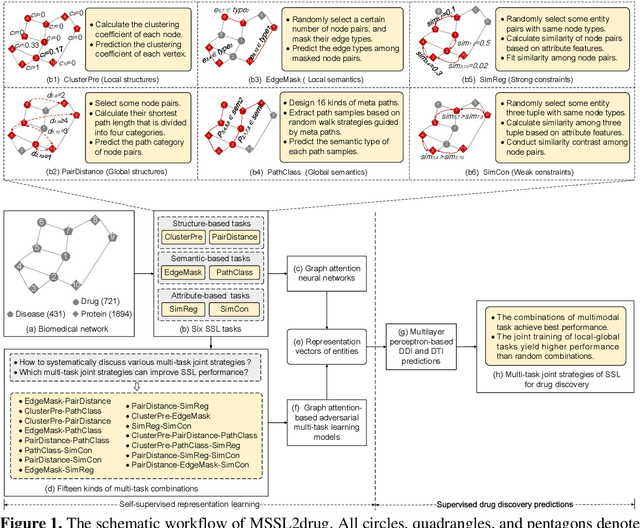

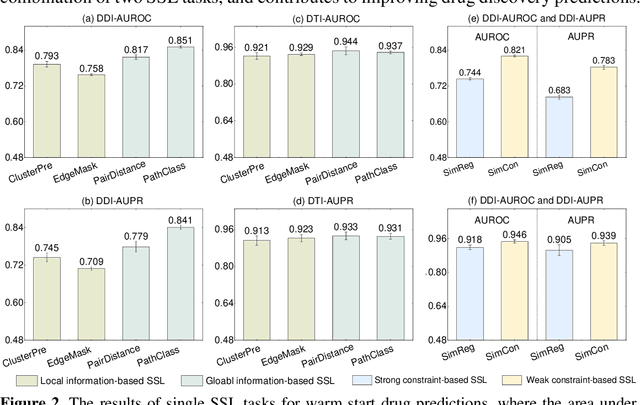
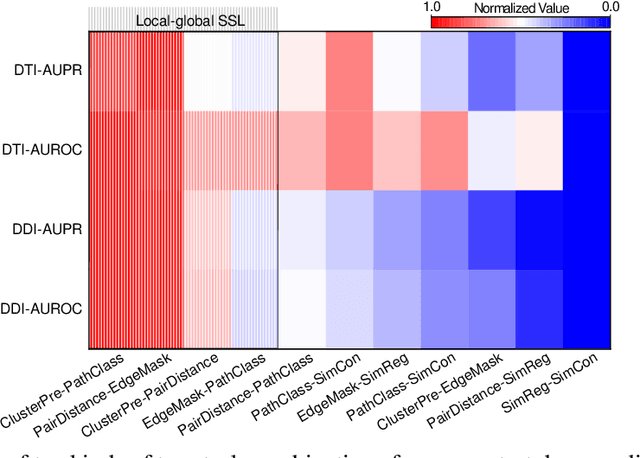
Self-supervised representation learning (SSL) on biomedical networks provides new opportunities for drug discovery which is lack of available biological or clinic phenotype. However, how to effectively combine multiple SSL models is challenging and rarely explored. Therefore, we propose multi-task joint strategies of self-supervised representation learning on biomedical networks for drug discovery, named MSSL2drug. We design six basic SSL tasks that are inspired by various modality features including structures, semantics, and attributes in biomedical heterogeneous networks. In addition, fifteen combinations of multiple tasks are evaluated by a graph attention-based adversarial multi-task learning framework in two drug discovery scenarios. The results suggest two important findings. (1) The combinations of multimodal tasks achieve the best performance compared to other multi-task joint strategies. (2) The joint training of local and global SSL tasks yields higher performance than random task combinations. Therefore, we conjecture that the multimodal and local-global combination strategies can be regarded as a guideline for multi-task SSL to drug discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge