Latent Patient Network Learning for Automatic Diagnosis
Paper and Code
Mar 27, 2020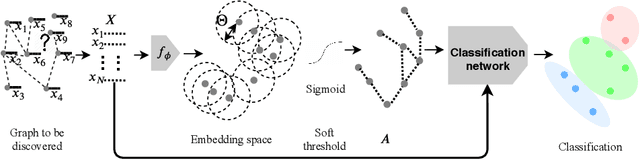
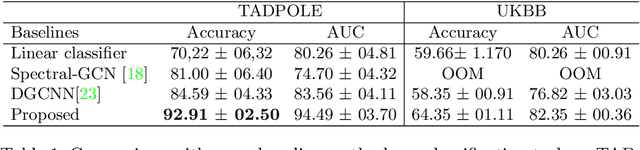
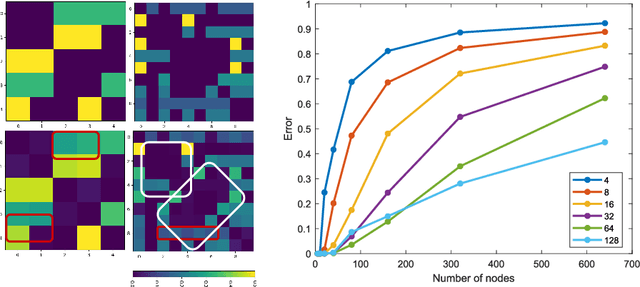
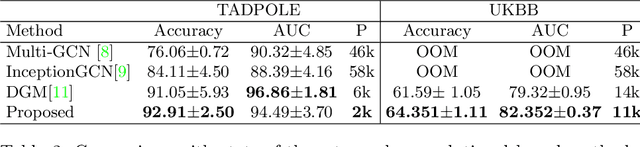
Recently, Graph Convolutional Networks (GCNs) has proven to be a powerful machine learning tool for Computer Aided Diagnosis (CADx) and disease prediction. A key component in these models is to build a population graph, where the graph adjacency matrix represents pair-wise patient similarities. Until now, the similarity metrics have been defined manually, usually based on meta-features like demographics or clinical scores. The definition of the metric, however, needs careful tuning, as GCNs are very sensitive to the graph structure. In this paper, we demonstrate for the first time in the CADx domain that it is possible to learn a single, optimal graph towards the GCN's downstream task of disease classification. To this end, we propose a novel, end-to-end trainable graph learning architecture for dynamic and localized graph pruning. Unlike commonly employed spectral GCN approaches, our GCN is spatial and inductive, and can thus infer previously unseen patients as well. We demonstrate significant classification improvements with our learned graph on two CADx problems in medicine. We further explain and visualize this result using an artificial dataset, underlining the importance of graph learning for more accurate and robust inference with GCNs in medical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge