Kernel classification of connectomes based on earth mover's distance between graph spectra
Paper and Code
Nov 27, 2016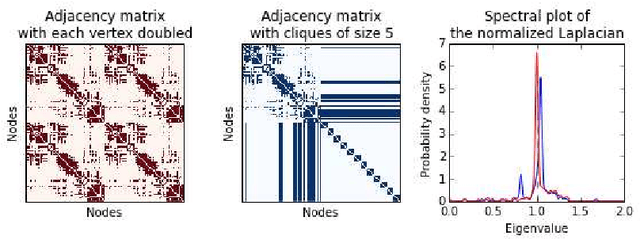
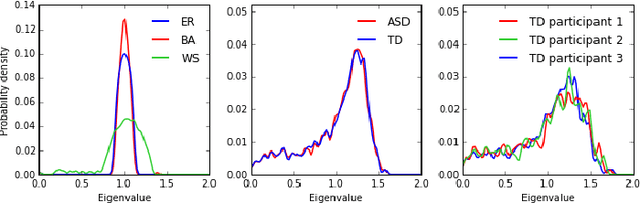
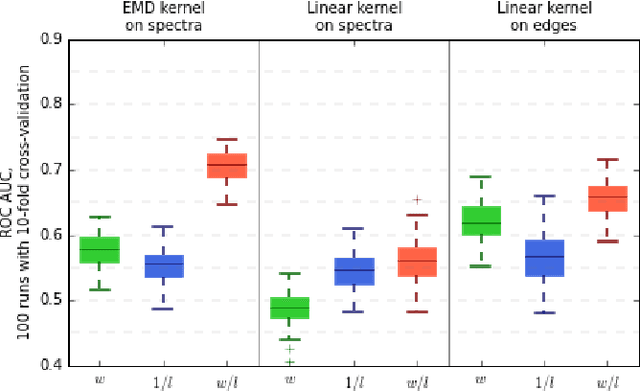
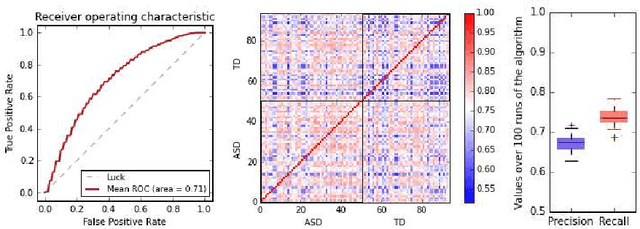
In this paper, we tackle a problem of predicting phenotypes from structural connectomes. We propose that normalized Laplacian spectra can capture structural properties of brain networks, and hence graph spectral distributions are useful for a task of connectome-based classification. We introduce a kernel that is based on earth mover's distance (EMD) between spectral distributions of brain networks. We access performance of an SVM classifier with the proposed kernel for a task of classification of autism spectrum disorder versus typical development based on a publicly available dataset. Classification quality (area under the ROC-curve) obtained with the EMD-based kernel on spectral distributions is 0.71, which is higher than that based on simpler graph embedding methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge