Incorporating Domain Knowledge into Medical NLI using Knowledge Graphs
Paper and Code
Aug 31, 2019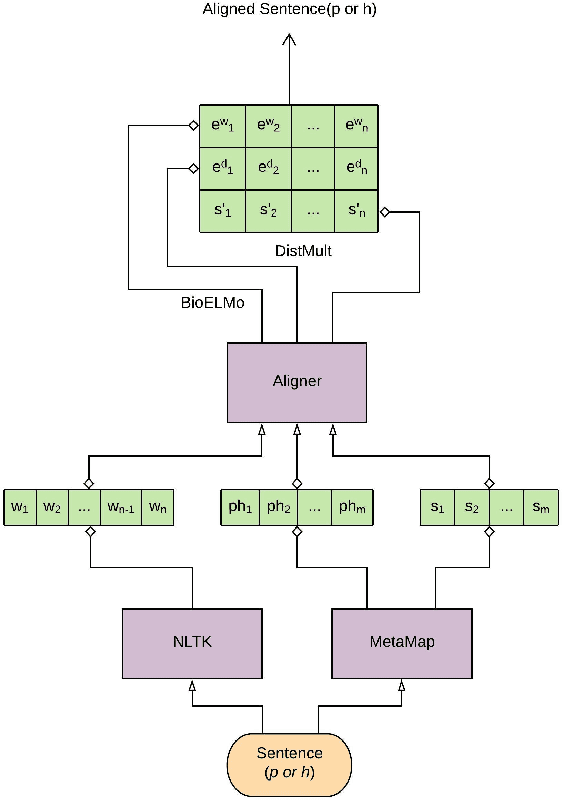
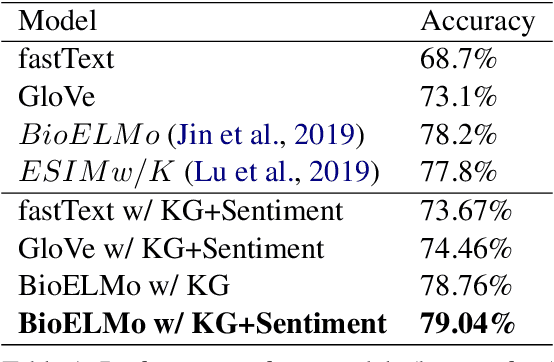
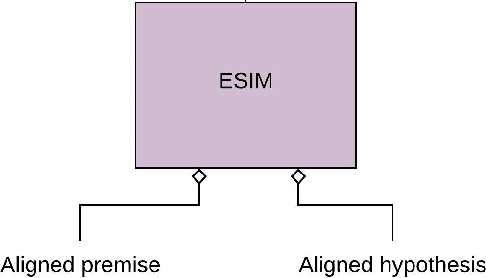
Recently, biomedical version of embeddings obtained from language models such as BioELMo have shown state-of-the-art results for the textual inference task in the medical domain. In this paper, we explore how to incorporate structured domain knowledge, available in the form of a knowledge graph (UMLS), for the Medical NLI task. Specifically, we experiment with fusing embeddings obtained from knowledge graph with the state-of-the-art approaches for NLI task (ESIM model). We also experiment with fusing the domain-specific sentiment information for the task. Experiments conducted on MedNLI dataset clearly show that this strategy improves the baseline BioELMo architecture for the Medical NLI task.
* EMNLP 2019 accepted short paper
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge