GV-Rep: A Large-Scale Dataset for Genetic Variant Representation Learning
Paper and Code
Jul 24, 2024


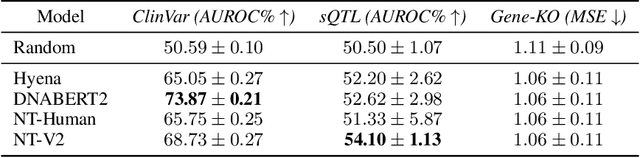
Genetic variants (GVs) are defined as differences in the DNA sequences among individuals and play a crucial role in diagnosing and treating genetic diseases. The rapid decrease in next generation sequencing cost has led to an exponential increase in patient-level GV data. This growth poses a challenge for clinicians who must efficiently prioritize patient-specific GVs and integrate them with existing genomic databases to inform patient management. To addressing the interpretation of GVs, genomic foundation models (GFMs) have emerged. However, these models lack standardized performance assessments, leading to considerable variability in model evaluations. This poses the question: How effectively do deep learning methods classify unknown GVs and align them with clinically-verified GVs? We argue that representation learning, which transforms raw data into meaningful feature spaces, is an effective approach for addressing both indexing and classification challenges. We introduce a large-scale Genetic Variant dataset, named GV-Rep, featuring variable-length contexts and detailed annotations, designed for deep learning models to learn GV representations across various traits, diseases, tissue types, and experimental contexts. Our contributions are three-fold: (i) Construction of a comprehensive dataset with 7 million records, each labeled with characteristics of the corresponding variants, alongside additional data from 17,548 gene knockout tests across 1,107 cell types, 1,808 variant combinations, and 156 unique clinically verified GVs from real-world patients. (ii) Analysis of the structure and properties of the dataset. (iii) Experimentation of the dataset with pre-trained GFMs. The results show a significant gap between GFMs current capabilities and accurate GV representation. We hope this dataset will help advance genomic deep learning to bridge this gap.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge