Generative Intervention Models for Causal Perturbation Modeling
Paper and Code
Nov 21, 2024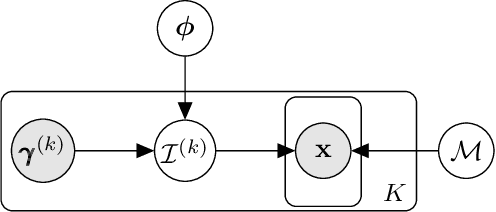
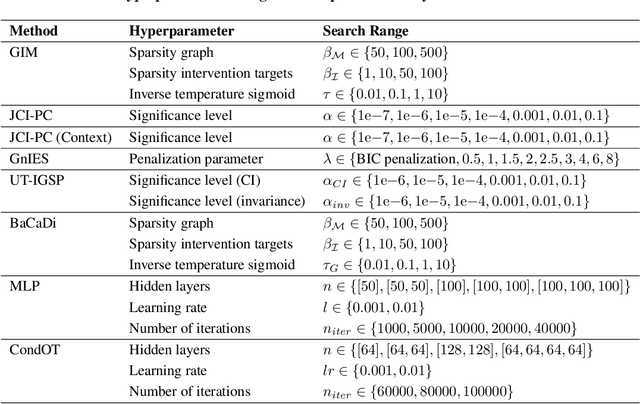
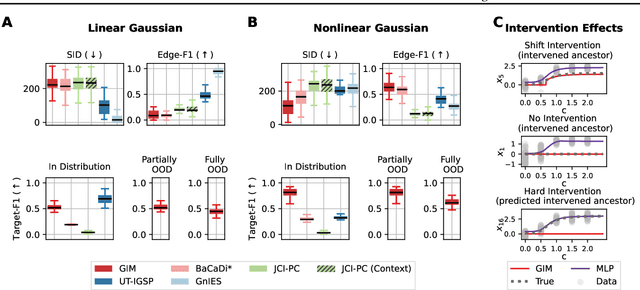
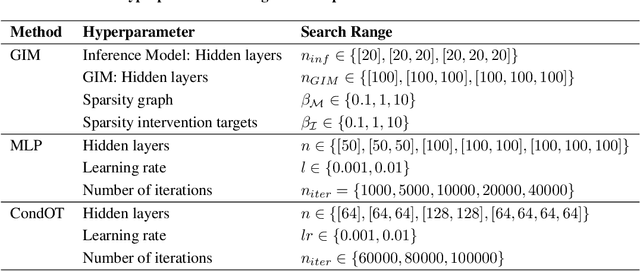
We consider the problem of predicting perturbation effects via causal models. In many applications, it is a priori unknown which mechanisms of a system are modified by an external perturbation, even though the features of the perturbation are available. For example, in genomics, some properties of a drug may be known, but not their causal effects on the regulatory pathways of cells. We propose a generative intervention model (GIM) that learns to map these perturbation features to distributions over atomic interventions in a jointly-estimated causal model. Contrary to prior approaches, this enables us to predict the distribution shifts of unseen perturbation features while gaining insights about their mechanistic effects in the underlying data-generating process. On synthetic data and scRNA-seq drug perturbation data, GIMs achieve robust out-of-distribution predictions on par with unstructured approaches, while effectively inferring the underlying perturbation mechanisms, often better than other causal inference methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge