Few-Shot 3D Volumetric Segmentation with Multi-Surrogate Fusion
Paper and Code
Aug 26, 2024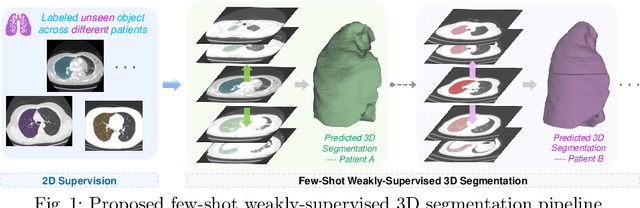
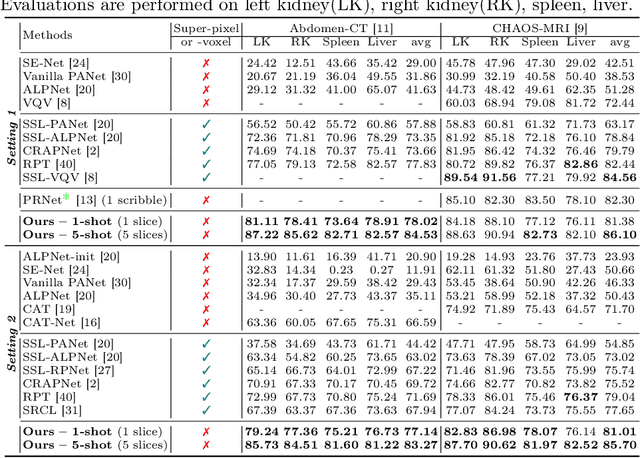
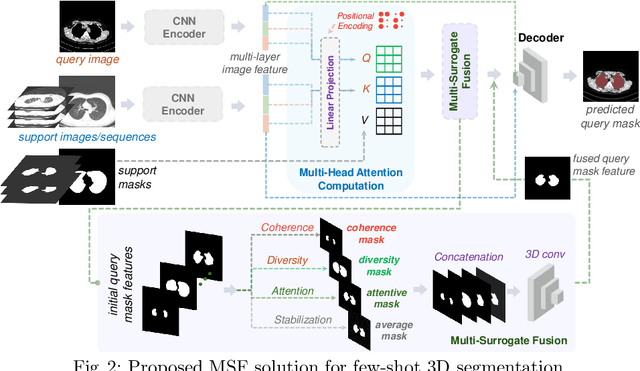
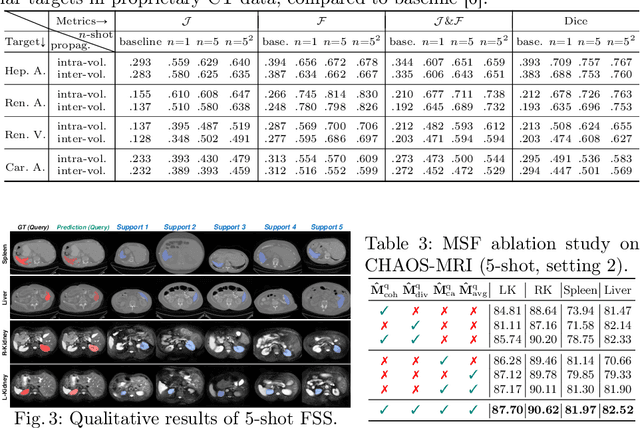
Conventional 3D medical image segmentation methods typically require learning heavy 3D networks (e.g., 3D-UNet), as well as large amounts of in-domain data with accurate pixel/voxel-level labels to avoid overfitting. These solutions are thus extremely time- and labor-expensive, but also may easily fail to generalize to unseen objects during training. To alleviate this issue, we present MSFSeg, a novel few-shot 3D segmentation framework with a lightweight multi-surrogate fusion (MSF). MSFSeg is able to automatically segment unseen 3D objects/organs (during training) provided with one or a few annotated 2D slices or 3D sequence segments, via learning dense query-support organ/lesion anatomy correlations across patient populations. Our proposed MSF module mines comprehensive and diversified morphology correlations between unlabeled and the few labeled slices/sequences through multiple designated surrogates, making it able to generate accurate cross-domain 3D segmentation masks given annotated slices or sequences. We demonstrate the effectiveness of our proposed framework by showing superior performance on conventional few-shot segmentation benchmarks compared to prior art, and remarkable cross-domain cross-volume segmentation performance on proprietary 3D segmentation datasets for challenging entities, i.e., tubular structures, with only limited 2D or 3D labels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge