EGFI: Drug-Drug Interaction Extraction and Generation with Fusion of Enriched Entity and Sentence Information
Paper and Code
Jan 25, 2021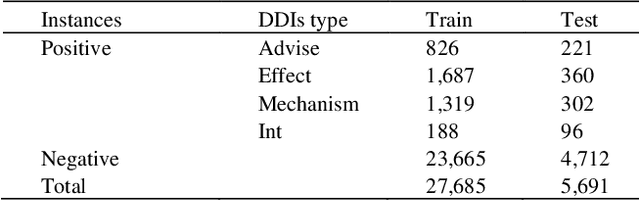
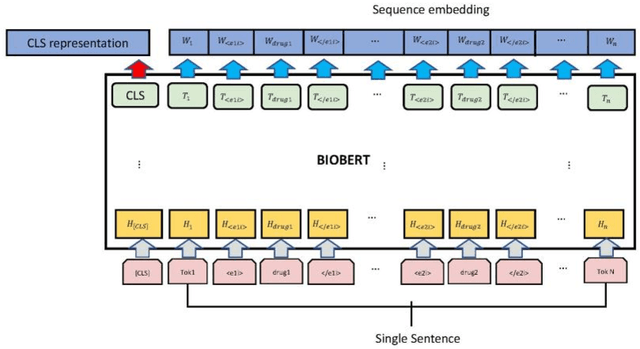
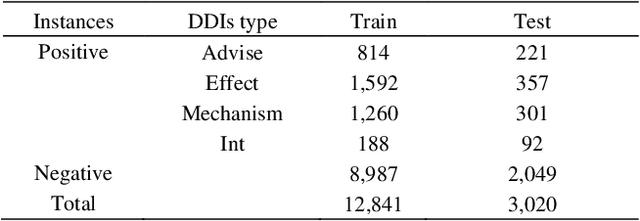
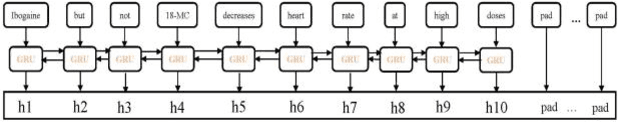
The rapid growth in literature accumulates diverse and yet comprehensive biomedical knowledge hidden to be mined such as drug interactions. However, it is difficult to extract the heterogeneous knowledge to retrieve or even discover the latest and novel knowledge in an efficient manner. To address such a problem, we propose EGFI for extracting and consolidating drug interactions from large-scale medical literature text data. Specifically, EGFI consists of two parts: classification and generation. In the classification part, EGFI encompasses the language model BioBERT which has been comprehensively pre-trained on biomedical corpus. In particular, we propose the multi-head attention mechanism and pack BiGRU to fuse multiple semantic information for rigorous context modeling. In the generation part, EGFI utilizes another pre-trained language model BioGPT-2 where the generation sentences are selected based on filtering rules. We evaluated the classification part on "DDIs 2013" dataset and "DTIs" dataset, achieving the FI score of 0.842 and 0.720 respectively. Moreover, we applied the classification part to distinguish high-quality generated sentences and verified with the exiting growth truth to confirm the filtered sentences. The generated sentences that are not recorded in DrugBank and DDIs 2013 dataset also demonstrate the potential of EGFI to identify novel drug relationships.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge