Efficient and Generic 1D Dilated Convolution Layer for Deep Learning
Paper and Code
Apr 16, 2021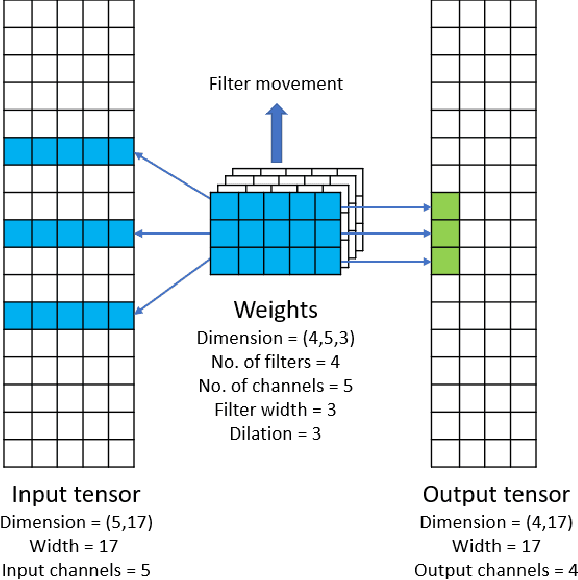
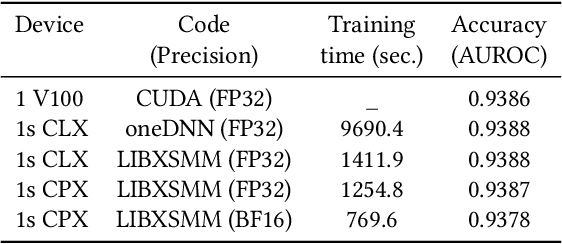
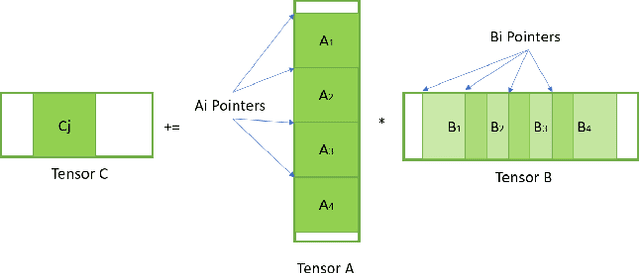
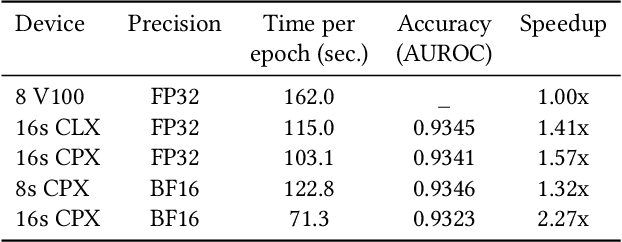
Convolutional neural networks (CNNs) have found many applications in tasks involving two-dimensional (2D) data, such as image classification and image processing. Therefore, 2D convolution layers have been heavily optimized on CPUs and GPUs. However, in many applications - for example genomics and speech recognition, the data can be one-dimensional (1D). Such applications can benefit from optimized 1D convolution layers. In this work, we introduce our efficient implementation of a generic 1D convolution layer covering a wide range of parameters. It is optimized for x86 CPU architectures, in particular, for architectures containing Intel AVX-512 and AVX-512 BFloat16 instructions. We use the LIBXSMM library's batch-reduce General Matrix Multiplication (BRGEMM) kernel for FP32 and BFloat16 precision. We demonstrate that our implementation can achieve up to 80% efficiency on Intel Xeon Cascade Lake and Cooper Lake CPUs. Additionally, we show the generalization capability of our BRGEMM based approach by achieving high efficiency across a range of parameters. We consistently achieve higher efficiency than the 1D convolution layer with Intel oneDNN library backend for varying input tensor widths, filter widths, number of channels, filters, and dilation parameters. Finally, we demonstrate the performance of our optimized 1D convolution layer by utilizing it in the end-to-end neural network training with real genomics datasets and achieve up to 6.86x speedup over the oneDNN library-based implementation on Cascade Lake CPUs. We also demonstrate the scaling with 16 sockets of Cascade/Cooper Lake CPUs and achieve significant speedup over eight V100 GPUs using a similar power envelop. In the end-to-end training, we get a speedup of 1.41x on Cascade Lake with FP32, 1.57x on Cooper Lake with FP32, and 2.27x on Cooper Lake with BFloat16 over eight V100 GPUs with FP32.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge