Discovering and Deciphering Relationships Across Disparate Data Modalities
Paper and Code
Sep 25, 2018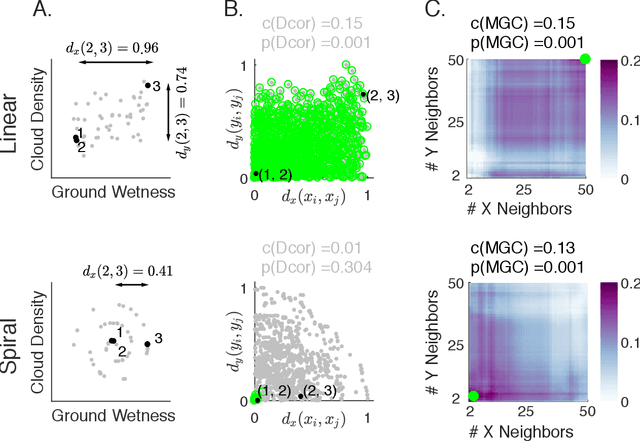
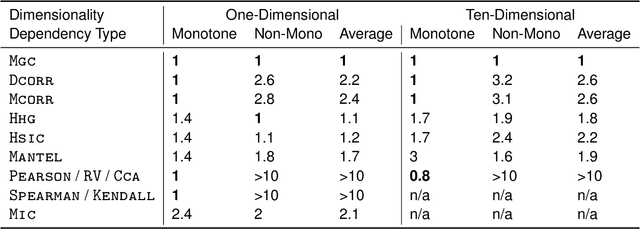
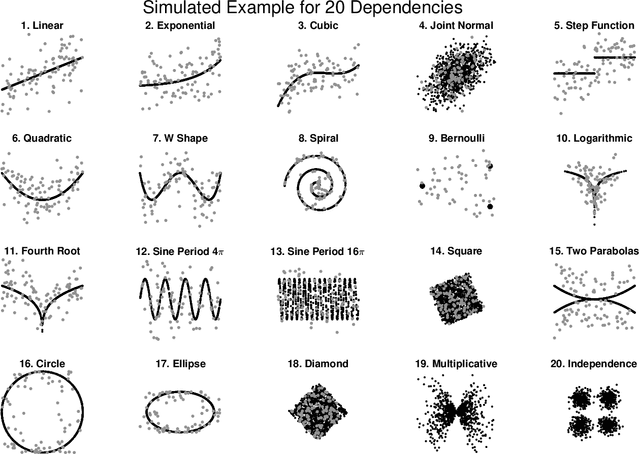
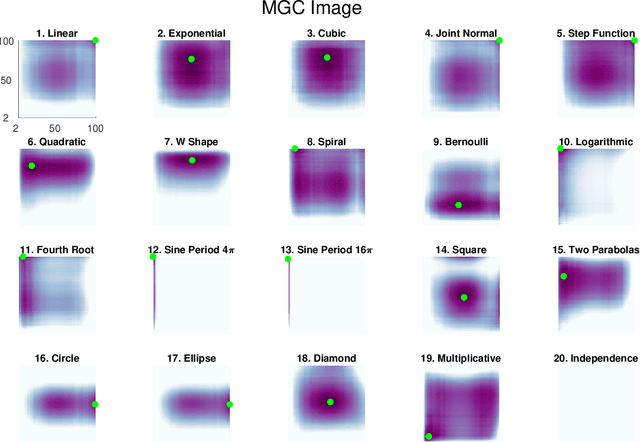
Understanding the relationships between different properties of data, such as whether a connectome or genome has information about disease status, is becoming increasingly important in modern biological datasets. While existing approaches can test whether two properties are related, they often require unfeasibly large sample sizes in real data scenarios, and do not provide any insight into how or why the procedure reached its decision. Our approach, "Multiscale Graph Correlation" (MGC), is a dependence test that juxtaposes previously disparate data science techniques, including k-nearest neighbors, kernel methods (such as support vector machines), and multiscale analysis (such as wavelets). Other methods typically require double or triple the number samples to achieve the same statistical power as MGC in a benchmark suite including high-dimensional and nonlinear relationships - spanning polynomial (linear, quadratic, cubic), trigonometric (sinusoidal, circular, ellipsoidal, spiral), geometric (square, diamond, W-shape), and other functions, with dimensionality ranging from 1 to 1000. Moreover, MGC uniquely provides a simple and elegant characterization of the potentially complex latent geometry underlying the relationship, providing insight while maintaining computational efficiency. In several real data applications, including brain imaging and cancer genetics, MGC is the only method that can both detect the presence of a dependency and provide specific guidance for the next experiment and/or analysis to conduct.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge