Differentiable Agent-based Epidemiology
Paper and Code
Jul 20, 2022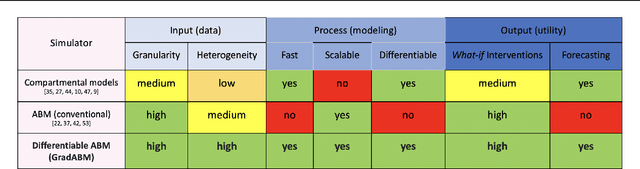
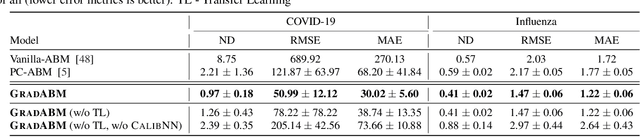
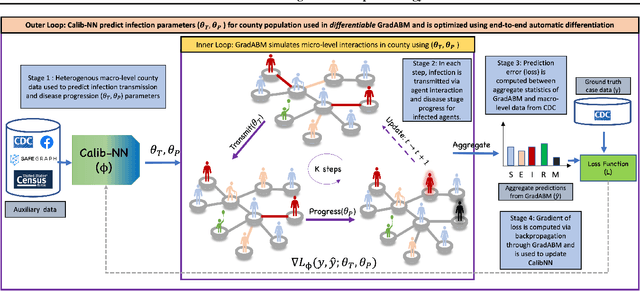

Mechanistic simulators are an indispensable tool for epidemiology to explore the behavior of complex, dynamic infections under varying conditions and navigate uncertain environments. ODE-based models are the dominant paradigm that enable fast simulations and are tractable to gradient-based optimization, but make simplifying assumptions about population homogeneity. Agent-based models (ABMs) are an increasingly popular alternative paradigm that can represent the heterogeneity of contact interactions with granular detail and agency of individual behavior. However, conventional ABM frameworks are not differentiable and present challenges in scalability; due to which it is non-trivial to connect them to auxiliary data sources easily. In this paper we introduce GradABM which is a new scalable, fast and differentiable design for ABMs. GradABM runs simulations in few seconds on commodity hardware and enables fast forward and differentiable inverse simulations. This makes it amenable to be merged with deep neural networks and seamlessly integrate heterogeneous data sources to help with calibration, forecasting and policy evaluation. We demonstrate the efficacy of GradABM via extensive experiments with real COVID-19 and influenza datasets. We are optimistic this work will bring ABM and AI communities closer together.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge