Deep Generative Modeling for Volume Reconstruction in Cryo-Electron Microscopy
Paper and Code
Jan 11, 2022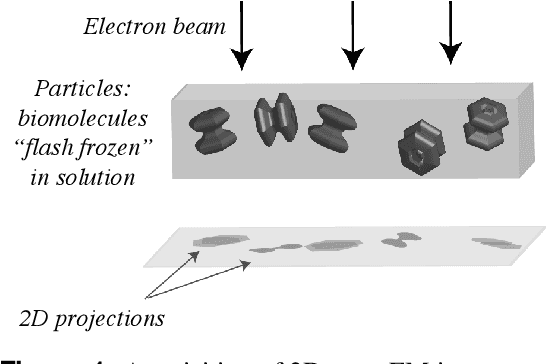

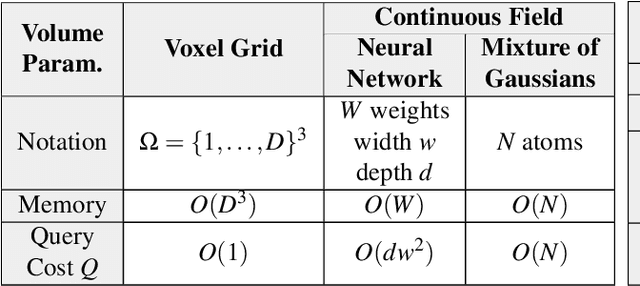
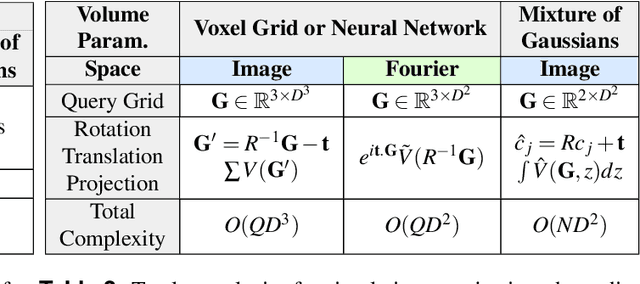
Recent breakthroughs in high resolution imaging of biomolecules in solution with cryo-electron microscopy (cryo-EM) have unlocked new doors for the reconstruction of molecular volumes, thereby promising further advances in biology, chemistry, and pharmacological research amongst others. Despite significant headway, the immense challenges in cryo-EM data analysis remain legion and intricately inter-disciplinary in nature, requiring insights from physicists, structural biologists, computer scientists, statisticians, and applied mathematicians. Meanwhile, recent next-generation volume reconstruction algorithms that combine generative modeling with end-to-end unsupervised deep learning techniques have shown promising results on simulated data, but still face considerable hurdles when applied to experimental cryo-EM images. In light of the proliferation of such methods and given the interdisciplinary nature of the task, we propose here a critical review of recent advances in the field of deep generative modeling for high resolution cryo-EM volume reconstruction. The present review aims to (i) compare and contrast these new methods, while (ii) presenting them from a perspective and using terminology familiar to scientists in each of the five aforementioned fields with no specific background in cryo-EM. The review begins with an introduction to the mathematical and computational challenges of deep generative models for cryo-EM volume reconstruction, along with an overview of the baseline methodology shared across this class of algorithms. Having established the common thread weaving through these different models, we provide a practical comparison of these state-of-the-art algorithms, highlighting their relative strengths and weaknesses, along with the assumptions that they rely on. This allows us to identify bottlenecks in current methods and avenues for future research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge