A Protein Structure Prediction Approach Leveraging Transformer and CNN Integration
Paper and Code
Mar 08, 2024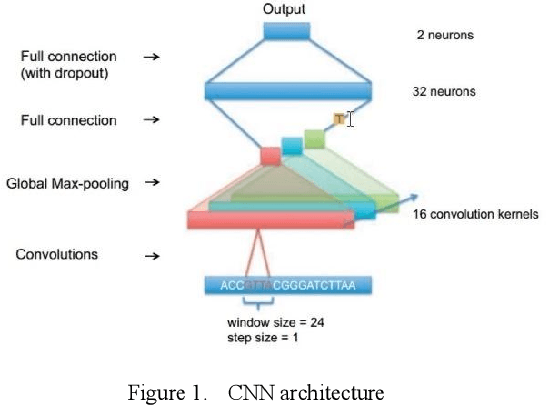
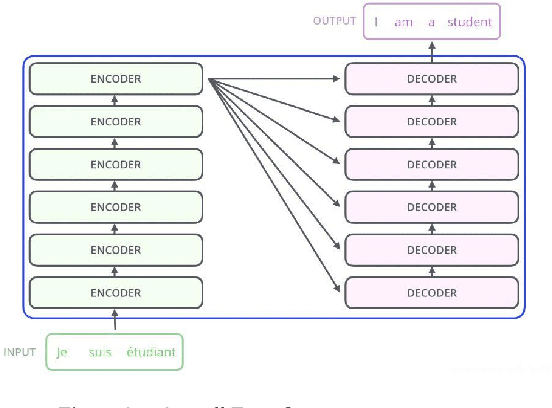
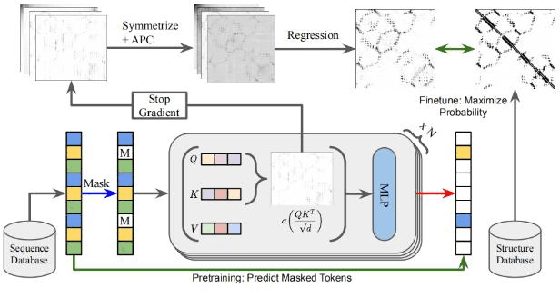
Proteins are essential for life, and their structure determines their function. The protein secondary structure is formed by the folding of the protein primary structure, and the protein tertiary structure is formed by the bending and folding of the secondary structure. Therefore, the study of protein secondary structure is very helpful to the overall understanding of protein structure. Although the accuracy of protein secondary structure prediction has continuously improved with the development of machine learning and deep learning, progress in the field of protein structure prediction, unfortunately, remains insufficient to meet the large demand for protein information. Therefore, based on the advantages of deep learning-based methods in feature extraction and learning ability, this paper adopts a two-dimensional fusion deep neural network model, DstruCCN, which uses Convolutional Neural Networks (CCN) and a supervised Transformer protein language model for single-sequence protein structure prediction. The training features of the two are combined to predict the protein Transformer binding site matrix, and then the three-dimensional structure is reconstructed using energy minimization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge