A Learning Search Algorithm for the Restricted Longest Common Subsequence Problem
Paper and Code
Oct 15, 2024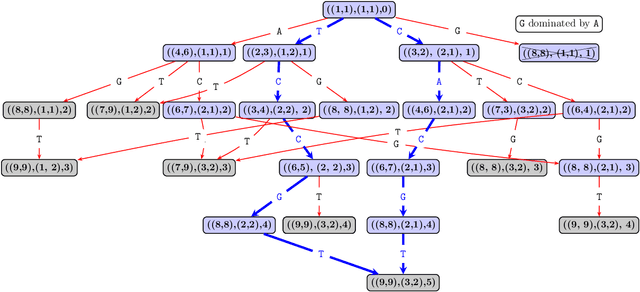
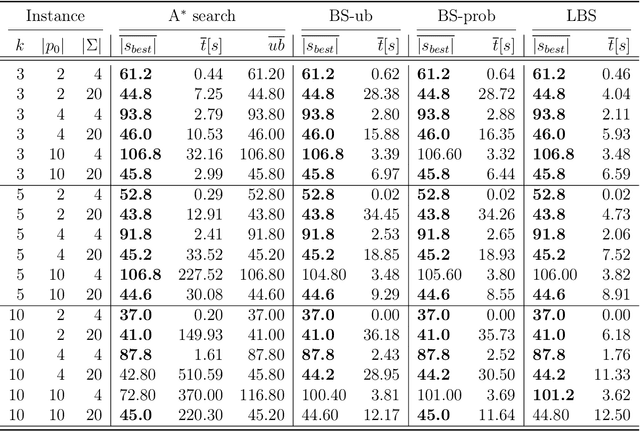
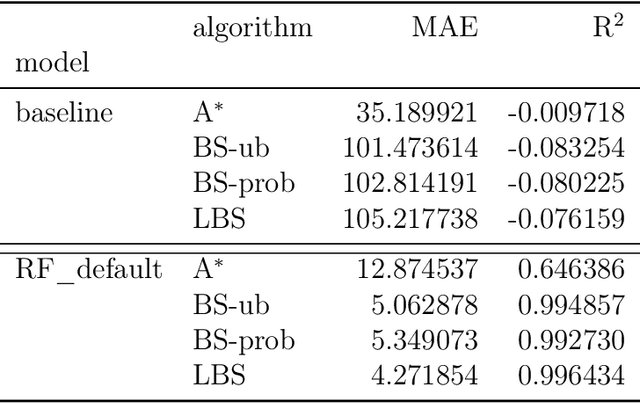
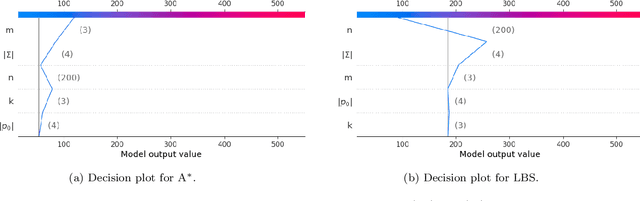
This paper addresses the Restricted Longest Common Subsequence (RLCS) problem, an extension of the well-known Longest Common Subsequence (LCS) problem. This problem has significant applications in bioinformatics, particularly for identifying similarities and discovering mutual patterns and important motifs among DNA, RNA, and protein sequences. Building on recent advancements in solving this problem through a general search framework, this paper introduces two novel heuristic approaches designed to enhance the search process by steering it towards promising regions in the search space. The first heuristic employs a probabilistic model to evaluate partial solutions during the search process. The second heuristic is based on a neural network model trained offline using a genetic algorithm. A key aspect of this approach is extracting problem-specific features of partial solutions and the complete problem instance. An effective hybrid method, referred to as the learning beam search, is developed by combining the trained neural network model with a beam search framework. An important contribution of this paper is found in the generation of real-world instances where scientific abstracts serve as input strings, and a set of frequently occurring academic words from the literature are used as restricted patterns. Comprehensive experimental evaluations demonstrate the effectiveness of the proposed approaches in solving the RLCS problem. Finally, an empirical explainability analysis is applied to the obtained results. In this way, key feature combinations and their respective contributions to the success or failure of the algorithms across different problem types are identified.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge