3D Reconstruction-Based Seed Counting of Sorghum Panicles for Agricultural Inspection
Paper and Code
Nov 14, 2022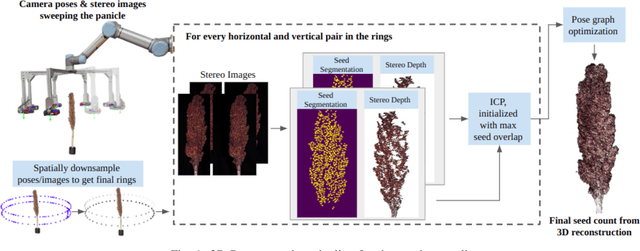

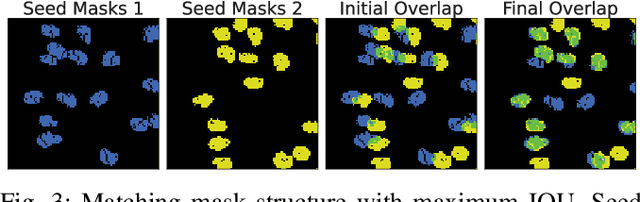
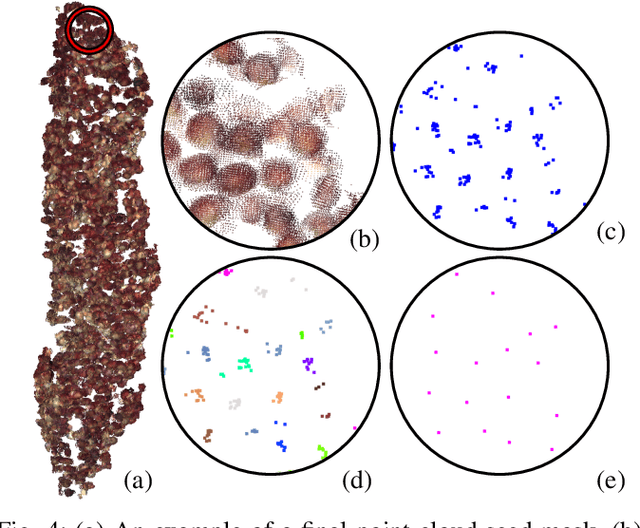
In this paper, we present a method for creating high-quality 3D models of sorghum panicles for phenotyping in breeding experiments. This is achieved with a novel reconstruction approach that uses seeds as semantic landmarks in both 2D and 3D. To evaluate the performance, we develop a new metric for assessing the quality of reconstructed point clouds without having a ground-truth point cloud. Finally, a counting method is presented where the density of seed centers in the 3D model allows 2D counts from multiple views to be effectively combined into a whole-panicle count. We demonstrate that using this method to estimate seed count and weight for sorghum outperforms count extrapolation from 2D images, an approach used in most state of the art methods for seeds and grains of comparable size.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge