Zhihong Ma
Generating high-quality 3DMPCs by adaptive data acquisition and NeREF-based reflectance correction to facilitate efficient plant phenotyping
May 11, 2023



Abstract:Non-destructive assessments of plant phenotypic traits using high-quality three-dimensional (3D) and multispectral data can deepen breeders' understanding of plant growth and allow them to make informed managerial decisions. However, subjective viewpoint selection and complex illumination effects under natural light conditions decrease the data quality and increase the difficulty of resolving phenotypic parameters. We proposed methods for adaptive data acquisition and reflectance correction respectively, to generate high-quality 3D multispectral point clouds (3DMPCs) of plants. In the first stage, we proposed an efficient next-best-view (NBV) planning method based on a novel UGV platform with a multi-sensor-equipped robotic arm. In the second stage, we eliminated the illumination effects by using the neural reference field (NeREF) to predict the digital number (DN) of the reference. We tested them on 6 perilla and 6 tomato plants, and selected 2 visible leaves and 4 regions of interest (ROIs) for each plant to assess the biomass and the chlorophyll content. For NBV planning, the average execution time for single perilla and tomato plant at a joint speed of 1.55 rad/s was 58.70 s and 53.60 s respectively. The whole-plant data integrity was improved by an average of 27% compared to using fixed viewpoints alone, and the coefficients of determination (R2) for leaf biomass estimation reached 0.99 and 0.92. For reflectance correction, the average root mean squared error of the reflectance spectra with hemisphere reference-based correction at different ROIs was 0.08 and 0.07 for perilla and tomato. The R2 of chlorophyll content estimation was 0.91 and 0.93 respectively when principal component analysis and Gaussian process regression were applied. Our approach is promising for generating high-quality 3DMPCs of plants under natural light conditions and facilitates accurate plant phenotyping.
PST: Plant Segmentation Transformer Enhanced Phenotyping of MLS Oilseed Rape Point Cloud
Jun 27, 2022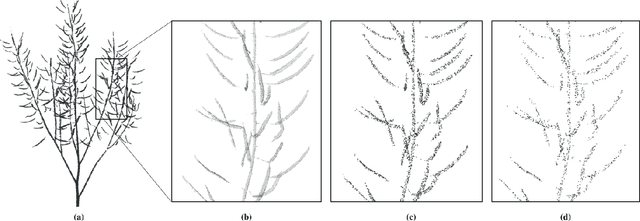


Abstract:Segmentation of plant point clouds to obtain high-precise morphological traits is essential for plant phenotyping and crop breeding. Although the bloom of deep learning methods has boosted much research on the segmentation of plant point cloud, most works follow the common practice of hard voxelization-based or down-sampling-based methods. They are limited to segmenting simple plant organs, overlooking the difficulties of resolving complex plant point clouds with high spatial resolution. In this study, we propose a deep learning network plant segmentation transformer (PST) to realize the semantic and instance segmentation of MLS (Mobile Laser Scanning) oilseed rape point cloud, which characterizes tiny siliques and dense points as the main traits targeted. PST is composed of: (i) a dynamic voxel feature encoder (DVFE) to aggregate per point features with raw spatial resolution; (ii) dual window sets attention block to capture the contextual information; (iii) a dense feature propagation module to obtain the final dense point feature map. The results proved that PST and PST-PointGroup (PG) achieved state-of-the-art performance in semantic and instance segmentation tasks. For semantic segmentation, PST reached 93.96%, 97.29%, 96.52%, 96.88%, and 97.07% in mean IoU, mean Precision, mean Recall, mean F1-score, and overall accuracy, respectively. For instance segmentation, PST-PG reached 89.51%, 89.85%, 88.83% and 82.53% in mCov, mWCov, mPerc90, and mRec90, respectively. This study extends the phenotyping of oilseed rape in an end-to-end way and proves that the deep learning method has a great potential for understanding dense plant point clouds with complex morphological traits.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge