Zhihang Dong
Adversarial and Safely Scaled Question Generation
Oct 17, 2022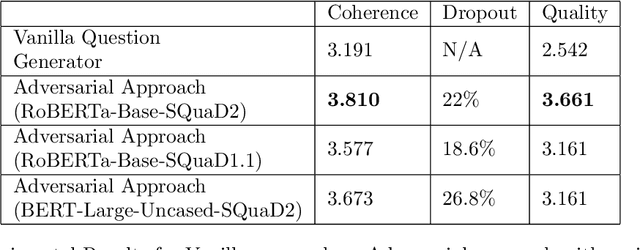
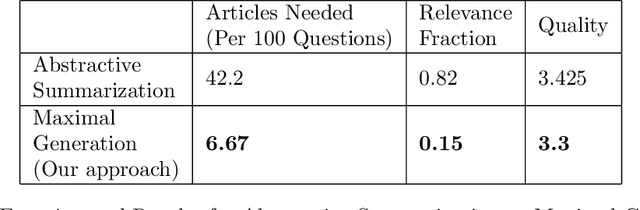
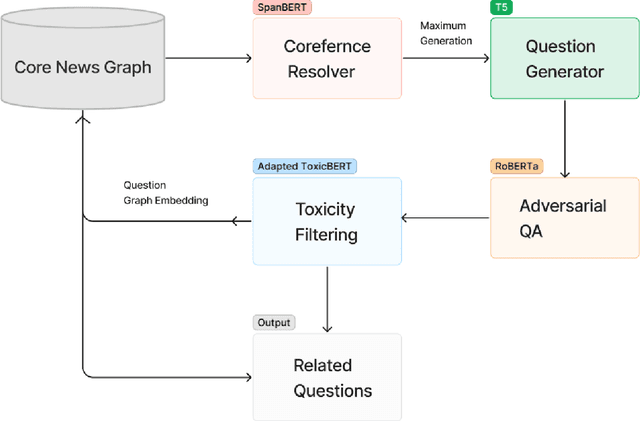
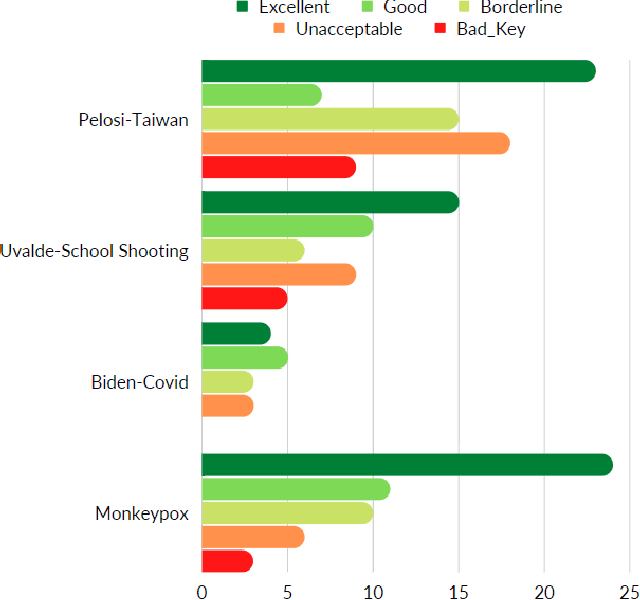
Abstract:Question generation has recently gained a lot of research interest, especially with the advent of large language models. In and of itself, question generation can be considered 'AI-hard', as there is a lack of unanimously agreed sense of what makes a question 'good' or 'bad'. In this paper, we tackle two fundamental problems in parallel: on one hand, we try to solve the scaling problem, where question-generation and answering applications have to be applied to a massive amount of text without ground truth labeling. The usual approach to solve this problem is to either downsample or summarize. However, there are critical risks of misinformation with these approaches. On the other hand, and related to the misinformation problem, we try to solve the 'safety' problem, as many public institutions rely on a much higher level of accuracy for the content they provide. We introduce an adversarial approach to tackle the question generation safety problem with scale. Specifically, we designed a question-answering system that specifically prunes out unanswerable questions that may be generated, and further increases the quality of the answers that are generated. We build a production-ready, easily-plugged pipeline that can be used on any given body of text, that is scalable and immune from generating any hate speech, profanity, or misinformation. Based on the results, we are able to generate more than six times the number of quality questions generated by the abstractive approach, with a perceived quality being 44% higher, according to a survey of 168 participants.
HMD-AMP: Protein Language-Powered Hierarchical Multi-label Deep Forest for Annotating Antimicrobial Peptides
Nov 11, 2021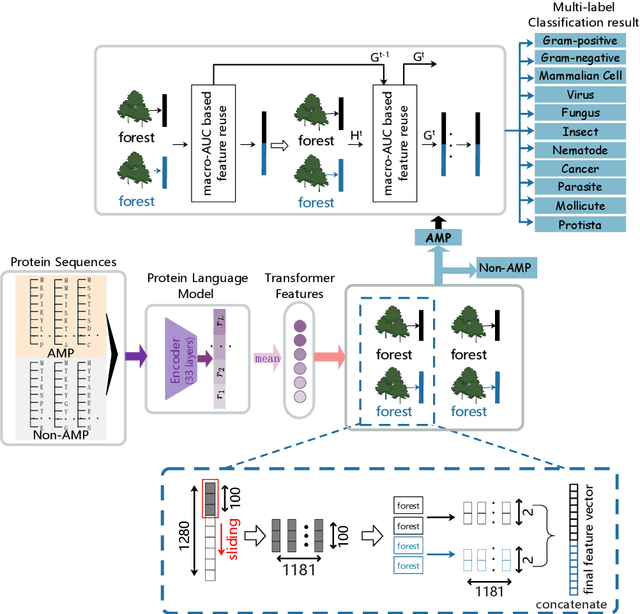
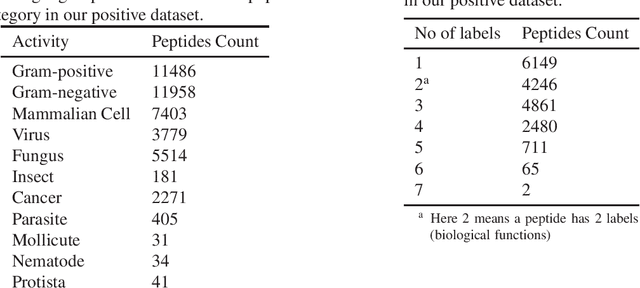
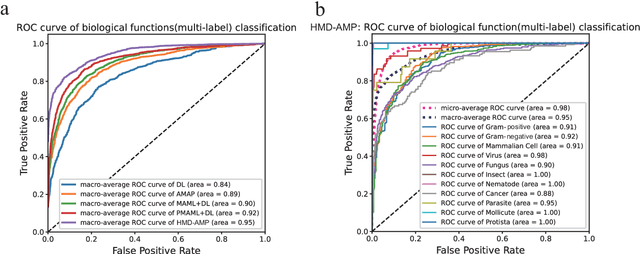
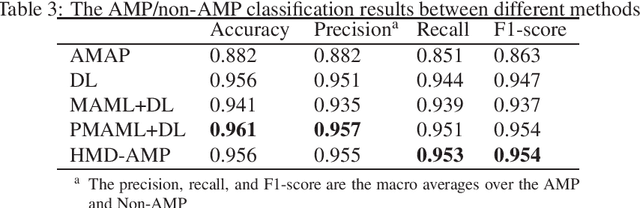
Abstract:Identifying the targets of an antimicrobial peptide is a fundamental step in studying the innate immune response and combating antibiotic resistance, and more broadly, precision medicine and public health. There have been extensive studies on the statistical and computational approaches to identify (i) whether a peptide is an antimicrobial peptide (AMP) or a non-AMP and (ii) which targets are these sequences effective to (Gram-positive, Gram-negative, etc.). Despite the existing deep learning methods on this problem, most of them are unable to handle the small AMP classes (anti-insect, anti-parasite, etc.). And more importantly, some AMPs can have multiple targets, which the previous methods fail to consider. In this study, we build a diverse and comprehensive multi-label protein sequence database by collecting and cleaning amino acids from various AMP databases. To generate efficient representations and features for the small classes dataset, we take advantage of a protein language model trained on 250 million protein sequences. Based on that, we develop an end-to-end hierarchical multi-label deep forest framework, HMD-AMP, to annotate AMP comprehensively. After identifying an AMP, it further predicts what targets the AMP can effectively kill from eleven available classes. Extensive experiments suggest that our framework outperforms state-of-the-art models in both the binary classification task and the multi-label classification task, especially on the minor classes.The model is robust against reduced features and small perturbations and produces promising results. We believe HMD-AMP contributes to both the future wet-lab investigations of the innate structural properties of different antimicrobial peptides and build promising empirical underpinnings for precise medicine with antibiotics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge