Zhifeng Kou
Empirical Evaluation of the Segment Anything Model (SAM) for Brain Tumor Segmentation
Oct 09, 2023
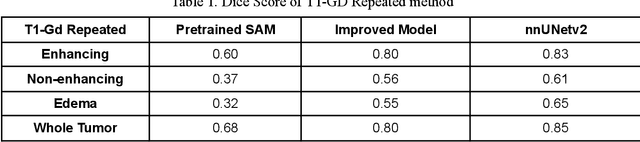

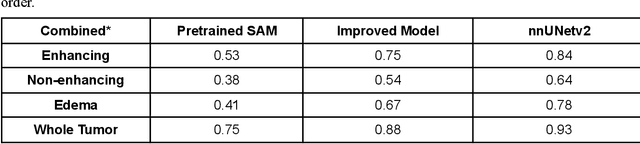
Abstract:Brain tumor segmentation presents a formidable challenge in the field of Medical Image Segmentation. While deep-learning models have been useful, human expert segmentation remains the most accurate method. The recently released Segment Anything Model (SAM) has opened up the opportunity to apply foundation models to this difficult task. However, SAM was primarily trained on diverse natural images. This makes applying SAM to biomedical segmentation, such as brain tumors with less defined boundaries, challenging. In this paper, we enhanced SAM's mask decoder using transfer learning with the Decathlon brain tumor dataset. We developed three methods to encapsulate the four-dimensional data into three dimensions for SAM. An on-the-fly data augmentation approach has been used with a combination of rotations and elastic deformations to increase the size of the training dataset. Two key metrics: the Dice Similarity Coefficient (DSC) and the Hausdorff Distance 95th Percentile (HD95), have been applied to assess the performance of our segmentation models. These metrics provided valuable insights into the quality of the segmentation results. In our evaluation, we compared this improved model to two benchmarks: the pretrained SAM and the widely used model, nnUNetv2. We find that the improved SAM shows considerable improvement over the pretrained SAM, while nnUNetv2 outperformed the improved SAM in terms of overall segmentation accuracy. Nevertheless, the improved SAM demonstrated slightly more consistent results than nnUNetv2, especially on challenging cases that can lead to larger Hausdorff distances. In the future, more advanced techniques can be applied in order to further improve the performance of SAM on brain tumor segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge