Zhenyu Yue
Controllable Edge-Type-Specific Interpretation in Multi-Relational Graph Neural Networks for Drug Response Prediction
Sep 03, 2024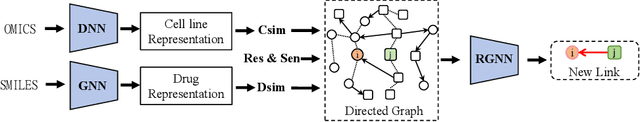
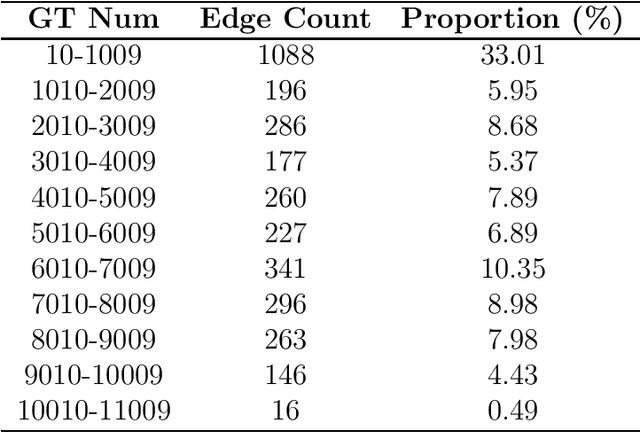
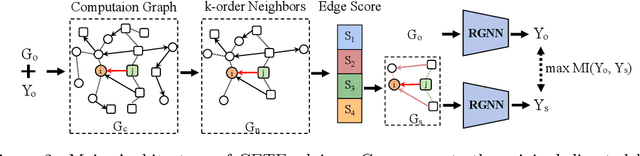
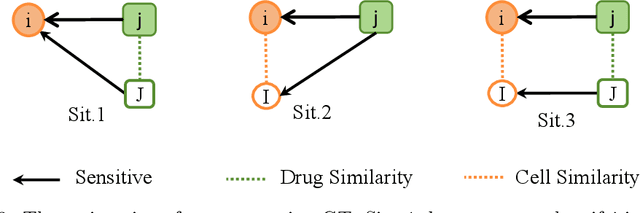
Abstract:Graph Neural Networks have been widely applied in critical decision-making areas that demand interpretable predictions, leading to the flourishing development of interpretability algorithms. However, current graph interpretability algorithms tend to emphasize generality and often overlook biological significance, thereby limiting their applicability in predicting cancer drug responses. In this paper, we propose a novel post-hoc interpretability algorithm for cancer drug response prediction, CETExplainer, which incorporates a controllable edge-type-specific weighting mechanism. It considers the mutual information between subgraphs and predictions, proposing a structural scoring approach to provide fine-grained, biologically meaningful explanations for predictive models. We also introduce a method for constructing ground truth based on real-world datasets to quantitatively evaluate the proposed interpretability algorithm. Empirical analysis on the real-world dataset demonstrates that CETExplainer achieves superior stability and improves explanation quality compared to leading algorithms, thereby offering a robust and insightful tool for cancer drug prediction.
DRExplainer: Quantifiable Interpretability in Drug Response Prediction with Directed Graph Convolutional Network
Aug 22, 2024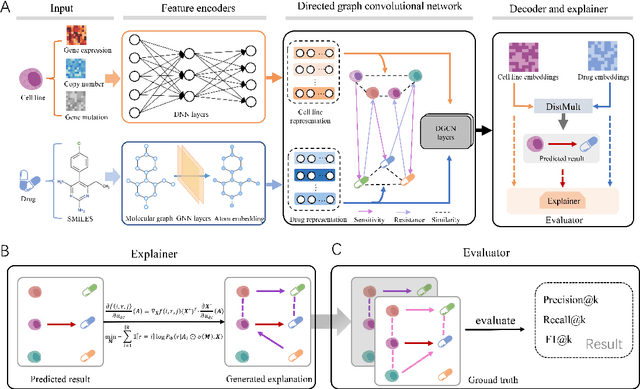
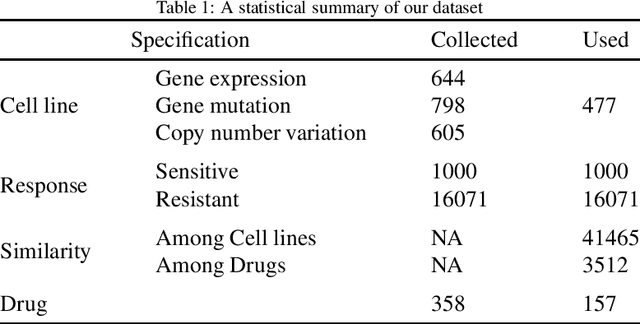
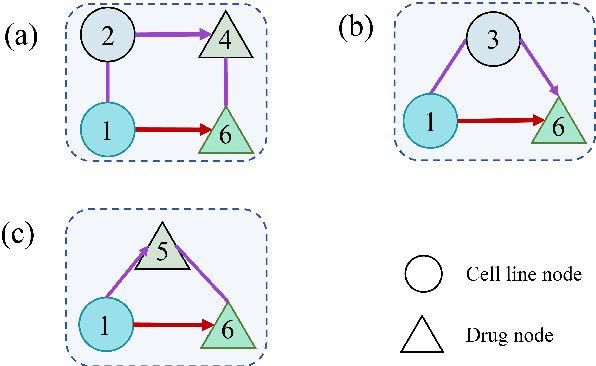

Abstract:Predicting the response of a cancer cell line to a therapeutic drug is pivotal for personalized medicine. Despite numerous deep learning methods that have been developed for drug response prediction, integrating diverse information about biological entities and predicting the directional response remain major challenges. Here, we propose a novel interpretable predictive model, DRExplainer, which leverages a directed graph convolutional network to enhance the prediction in a directed bipartite network framework. DRExplainer constructs a directed bipartite network integrating multi-omics profiles of cell lines, the chemical structure of drugs and known drug response to achieve directed prediction. Then, DRExplainer identifies the most relevant subgraph to each prediction in this directed bipartite network by learning a mask, facilitating critical medical decision-making. Additionally, we introduce a quantifiable method for model interpretability that leverages a ground truth benchmark dataset curated from biological features. In computational experiments, DRExplainer outperforms state-of-the-art predictive methods and another graph-based explanation method under the same experimental setting. Finally, the case studies further validate the interpretability and the effectiveness of DRExplainer in predictive novel drug response. Our code is available at: https://github.com/vshy-dream/DRExplainer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge