Yousuf Babiker M. Osman
Automating Vessel Segmentation in the Heart and Brain: A Trend to Develop Multi-Modality and Label-Efficient Deep Learning Techniques
Apr 02, 2024



Abstract:Cardio-cerebrovascular diseases are the leading causes of mortality worldwide, whose accurate blood vessel segmentation is significant for both scientific research and clinical usage. However, segmenting cardio-cerebrovascular structures from medical images is very challenging due to the presence of thin or blurred vascular shapes, imbalanced distribution of vessel and non-vessel pixels, and interference from imaging artifacts. These difficulties make manual or semi-manual segmentation methods highly time-consuming, labor-intensive, and prone to errors with interobserver variability, where different experts may produce different segmentations from a variety of modalities. Consequently, there is a growing interest in developing automated algorithms. This paper provides an up-to-date survey of deep learning techniques, for cardio-cerebrovascular segmentation. It analyzes the research landscape, surveys recent approaches, and discusses challenges such as the scarcity of accurately annotated data and variability. This paper also illustrates the urgent needs for developing multi-modality label-efficient deep learning techniques. To the best of our knowledge, this paper is the first comprehensive survey of deep learning approaches that effectively segment vessels in both the heart and brain. It aims to advance automated segmentation techniques for cardio-cerebrovascular diseases, benefiting researchers and healthcare professionals.
Collaborative Learning for Annotation-Efficient Volumetric MR Image Segmentation
Dec 18, 2023



Abstract:Background: Deep learning has presented great potential in accurate MR image segmentation when enough labeled data are provided for network optimization. However, manually annotating 3D MR images is tedious and time-consuming, requiring experts with rich domain knowledge and experience. Purpose: To build a deep learning method exploring sparse annotations, namely only a single 2D slice label for each 3D training MR image. Population: 3D MR images of 150 subjects from two publicly available datasets were included. Among them, 50 (1,377 image slices) are for prostate segmentation. The other 100 (8,800 image slices) are for left atrium segmentation. Five-fold cross-validation experiments were carried out utilizing the first dataset. For the second dataset, 80 subjects were used for training and 20 were used for testing. Assessment: A collaborative learning method by integrating the strengths of semi-supervised and self-supervised learning schemes was developed. The method was trained using labeled central slices and unlabeled non-central slices. Segmentation performance on testing set was reported quantitatively and qualitatively. Results: Compared to FS-LCS, MT, UA-MT, DCT-Seg, ICT, and AC-MT, the proposed method achieved a substantial improvement in segmentation accuracy, increasing the mean B-IoU significantly by more than 10.0% for prostate segmentation (proposed method B-IoU: 70.3% vs. ICT B-IoU: 60.3%) and by more than 6.0% for left atrium segmentation (proposed method B-IoU: 66.1% vs. ICT B-IoU: 60.1%).
Uncertainty-Aware Multi-Parametric Magnetic Resonance Image Information Fusion for 3D Object Segmentation
Nov 16, 2022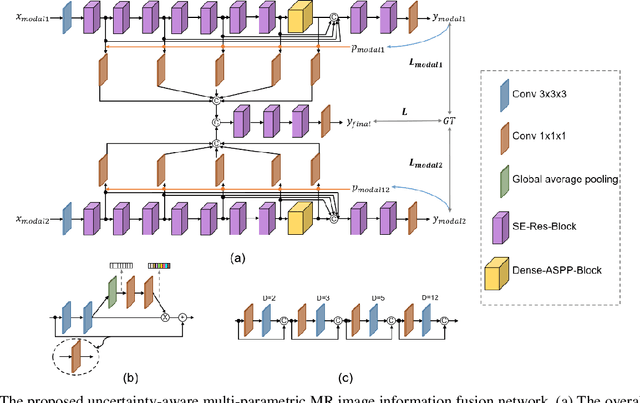
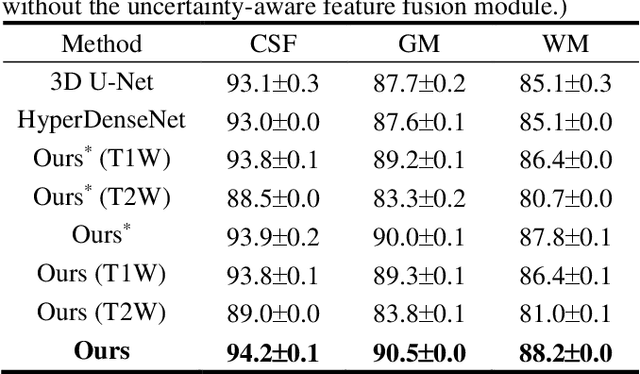
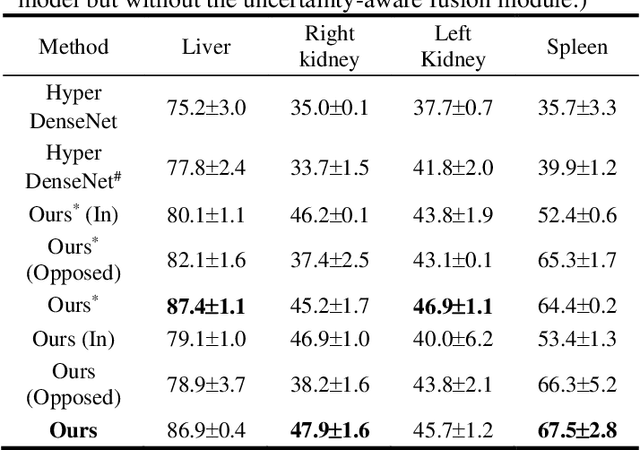
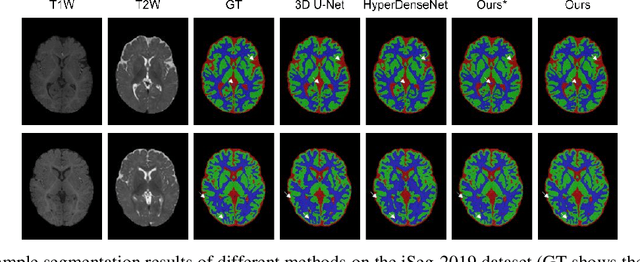
Abstract:Multi-parametric magnetic resonance (MR) imaging is an indispensable tool in the clinic. Consequently, automatic volume-of-interest segmentation based on multi-parametric MR imaging is crucial for computer-aided disease diagnosis, treatment planning, and prognosis monitoring. Despite the extensive studies conducted in deep learning-based medical image analysis, further investigations are still required to effectively exploit the information provided by different imaging parameters. How to fuse the information is a key question in this field. Here, we propose an uncertainty-aware multi-parametric MR image feature fusion method to fully exploit the information for enhanced 3D image segmentation. Uncertainties in the independent predictions of individual modalities are utilized to guide the fusion of multi-modal image features. Extensive experiments on two datasets, one for brain tissue segmentation and the other for abdominal multi-organ segmentation, have been conducted, and our proposed method achieves better segmentation performance when compared to existing models.
Semi-Supervised and Self-Supervised Collaborative Learning for Prostate 3D MR Image Segmentation
Nov 16, 2022


Abstract:Volumetric magnetic resonance (MR) image segmentation plays an important role in many clinical applications. Deep learning (DL) has recently achieved state-of-the-art or even human-level performance on various image segmentation tasks. Nevertheless, manually annotating volumetric MR images for DL model training is labor-exhaustive and time-consuming. In this work, we aim to train a semi-supervised and self-supervised collaborative learning framework for prostate 3D MR image segmentation while using extremely sparse annotations, for which the ground truth annotations are provided for just the central slice of each volumetric MR image. Specifically, semi-supervised learning and self-supervised learning methods are used to generate two independent sets of pseudo labels. These pseudo labels are then fused by Boolean operation to extract a more confident pseudo label set. The images with either manual or network self-generated labels are then employed to train a segmentation model for target volume extraction. Experimental results on a publicly available prostate MR image dataset demonstrate that, while requiring significantly less annotation effort, our framework generates very encouraging segmentation results. The proposed framework is very useful in clinical applications when training data with dense annotations are difficult to obtain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge