Yosuke Toda
Zero-shot Hierarchical Plant Segmentation via Foundation Segmentation Models and Text-to-image Attention
Sep 11, 2025Abstract:Foundation segmentation models achieve reasonable leaf instance extraction from top-view crop images without training (i.e., zero-shot). However, segmenting entire plant individuals with each consisting of multiple overlapping leaves remains challenging. This problem is referred to as a hierarchical segmentation task, typically requiring annotated training datasets, which are often species-specific and require notable human labor. To address this, we introduce ZeroPlantSeg, a zero-shot segmentation for rosette-shaped plant individuals from top-view images. We integrate a foundation segmentation model, extracting leaf instances, and a vision-language model, reasoning about plants' structures to extract plant individuals without additional training. Evaluations on datasets with multiple plant species, growth stages, and shooting environments demonstrate that our method surpasses existing zero-shot methods and achieves better cross-domain performance than supervised methods. Implementations are available at https://github.com/JunhaoXing/ZeroPlantSeg.
TreeFormer: Single-view Plant Skeleton Estimation via Tree-constrained Graph Generation
Nov 25, 2024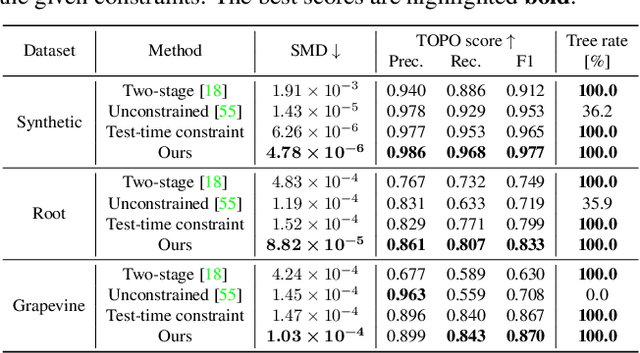
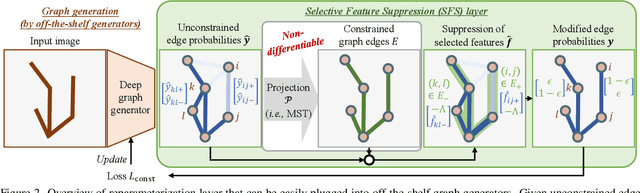
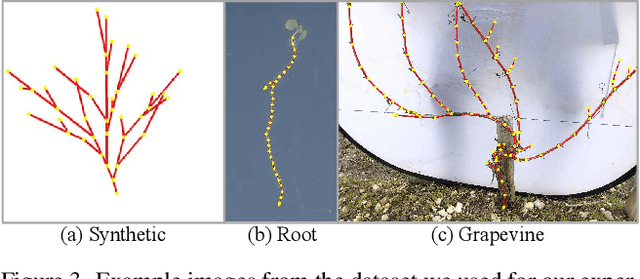
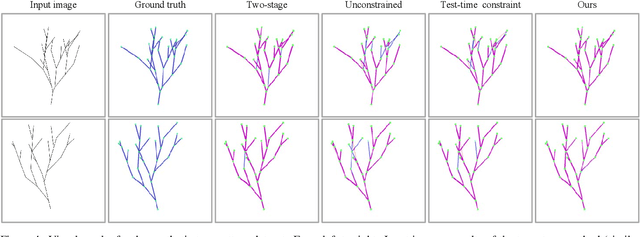
Abstract:Accurate estimation of plant skeletal structure (e.g., branching structure) from images is essential for smart agriculture and plant science. Unlike human skeletons with fixed topology, plant skeleton estimation presents a unique challenge, i.e., estimating arbitrary tree graphs from images. While recent graph generation methods successfully infer thin structures from images, it is challenging to constrain the output graph strictly to a tree structure. To this problem, we present TreeFormer, a plant skeleton estimator via tree-constrained graph generation. Our approach combines learning-based graph generation with traditional graph algorithms to impose the constraints during the training loop. Specifically, our method projects an unconstrained graph onto a minimum spanning tree (MST) during the training loop and incorporates this prior knowledge into the gradient descent optimization by suppressing unwanted feature values. Experiments show that our method accurately estimates target plant skeletal structures for multiple domains: Synthetic tree patterns, real botanical roots, and grapevine branches. Our implementations are available at https://github.com/huntorochi/TreeFormer/.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge