Yongxiang Wang
PETformer: Long-term Time Series Forecasting via Placeholder-enhanced Transformer
Aug 09, 2023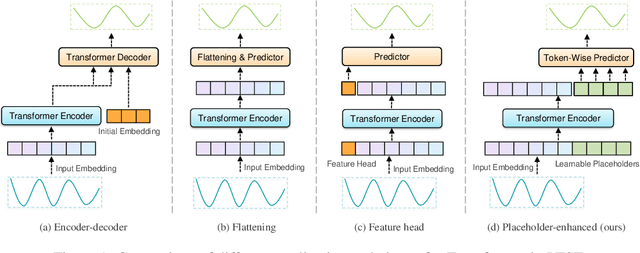

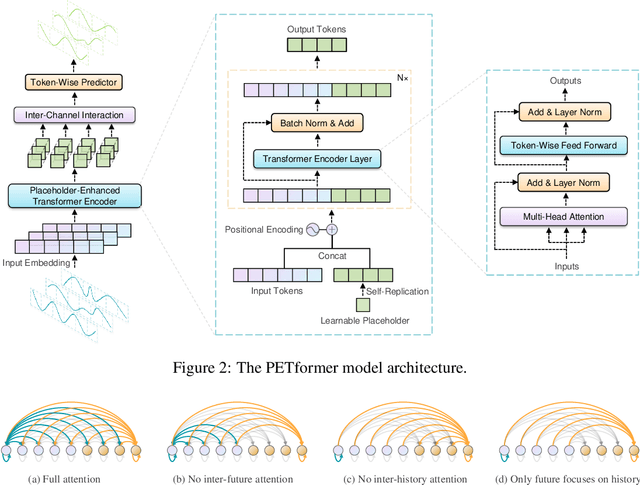
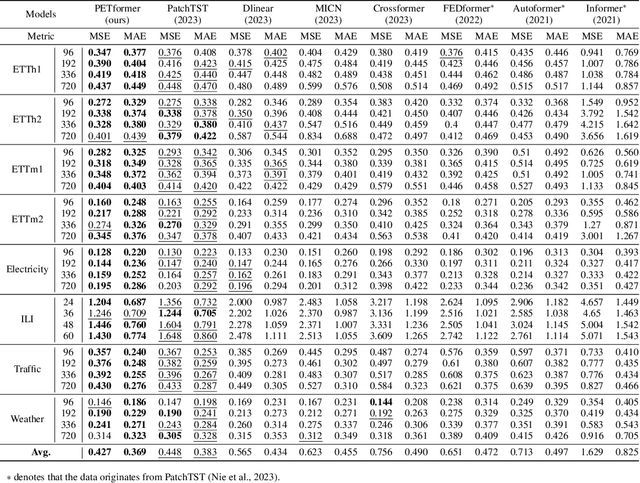
Abstract:Recently, Transformer-based models have shown remarkable performance in long-term time series forecasting (LTSF) tasks due to their ability to model long-term dependencies. However, the validity of Transformers for LTSF tasks remains debatable, particularly since recent work has shown that simple linear models can outperform numerous Transformer-based approaches. This suggests that there are limitations to the application of Transformer in LTSF. Therefore, this paper investigates three key issues when applying Transformer to LTSF: temporal continuity, information density, and multi-channel relationships. Accordingly, we propose three innovative solutions, including Placeholder Enhancement Technique (PET), Long Sub-sequence Division (LSD), and Multi-channel Separation and Interaction (MSI), which together form a novel model called PETformer. These three key designs introduce prior biases suitable for LTSF tasks. Extensive experiments have demonstrated that PETformer achieves state-of-the-art (SOTA) performance on eight commonly used public datasets for LTSF, outperforming all other models currently available. This demonstrates that Transformer still possesses powerful capabilities in LTSF.
Efficient online learning for large-scale peptide identification
May 08, 2018



Abstract:Motivation: Post-database searching is a key procedure in peptide dentification with tandem mass spectrometry (MS/MS) strategies for refining peptide-spectrum matches (PSMs) generated by database search engines. Although many statistical and machine learning-based methods have been developed to improve the accuracy of peptide identification, the challenge remains on large-scale datasets and datasets with an extremely large proportion of false positives (hard datasets). A more efficient learning strategy is required for improving the performance of peptide identification on challenging datasets. Results: In this work, we present an online learning method to conquer the challenges remained for exiting peptide identification algorithms. We propose a cost-sensitive learning model by using different loss functions for decoy and target PSMs respectively. A larger penalty for wrongly selecting decoy PSMs than that for target PSMs, and thus the new model can reduce its false discovery rate on hard datasets. Also, we design an online learning algorithm, OLCS-Ranker, to solve the proposed learning model. Rather than taking all training data samples all at once, OLCS-Ranker iteratively feeds in only one training sample into the learning model at each round. As a result, the memory requirement is significantly reduced for large-scale problems. Experimental studies show that OLCS-Ranker outperforms benchmark methods, such as CRanker and Batch-CS-Ranker, in terms of accuracy and stability. Furthermore, OLCS-Ranker is 15--85 times faster than CRanker method on large datasets. Availability and implementation: OLCS-Ranker software is available at no charge for non-commercial use at https://github.com/Isaac-QiXing/CRanker.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge