Yefeng Wang
Deep Learning Models in Detection of Dietary Supplement Adverse Event Signals from Twitter
Jun 21, 2021
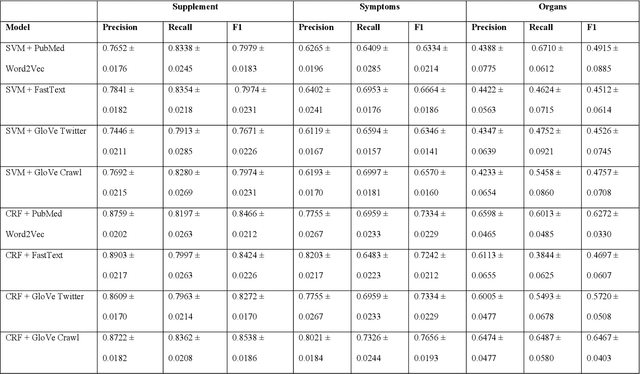

Abstract:Objective: The objective of this study is to develop a deep learning pipeline to detect signals on dietary supplement-related adverse events (DS AEs) from Twitter. Material and Methods: We obtained 247,807 tweets ranging from 2012 to 2018 that mentioned both DS and AE. We annotated biomedical entities and relations on 2,000 randomly selected tweets. For the concept extraction task, we compared the performance of traditional word embeddings with SVM, CRF and LSTM-CRF classifiers to BERT models. For the relation extraction task, we compared GloVe vectors with CNN classifiers to BERT models. We chose the best performing models in each task to assemble an end-to-end deep learning pipeline to detect DS AE signals and compared the results to the known DS AEs from a DS knowledge base (i.e., iDISK). Results: In both tasks, the BERT-based models outperformed traditional word embeddings. The best performing concept extraction model is the BioBERT model that can identify supplement, symptom, and body organ entities with F1-scores of 0.8646, 0.8497, and 0.7104, respectively. The best performing relation extraction model is the BERT model that can identify purpose and AE relations with F1-scores of 0.8335 and 0.7538, respectively. The end-to-end pipeline was able to extract DS indication and DS AEs with an F1-score of 0.7459 and 0,7414, respectively. Comparing to the iDISK, we could find both known and novel DS-AEs. Conclusion: We have demonstrated the feasibility of detecting DS AE signals from Twitter with a BioBERT-based deep learning pipeline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge