Yeeleng Scott Vang
Deep Multi-instance Networks with Sparse Label Assignment for Whole Mammogram Classification
May 23, 2017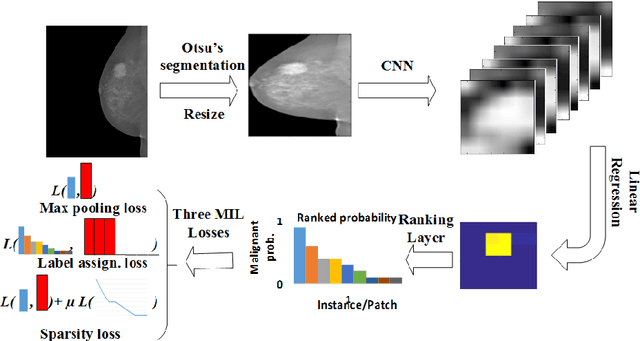
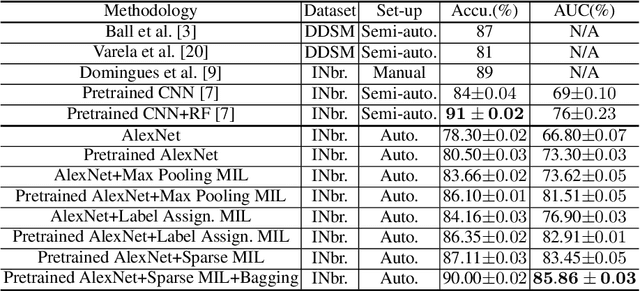
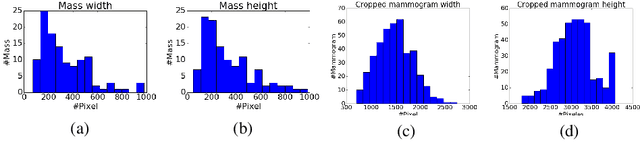
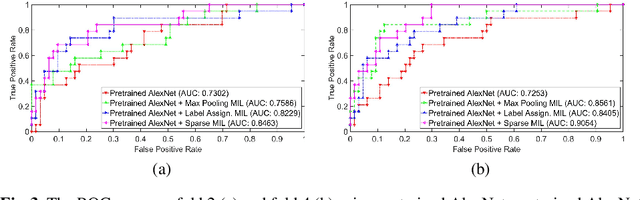
Abstract:Mammogram classification is directly related to computer-aided diagnosis of breast cancer. Traditional methods rely on regions of interest (ROIs) which require great efforts to annotate. Inspired by the success of using deep convolutional features for natural image analysis and multi-instance learning (MIL) for labeling a set of instances/patches, we propose end-to-end trained deep multi-instance networks for mass classification based on whole mammogram without the aforementioned ROIs. We explore three different schemes to construct deep multi-instance networks for whole mammogram classification. Experimental results on the INbreast dataset demonstrate the robustness of proposed networks compared to previous work using segmentation and detection annotations.
HLA class I binding prediction via convolutional neural networks
Apr 12, 2017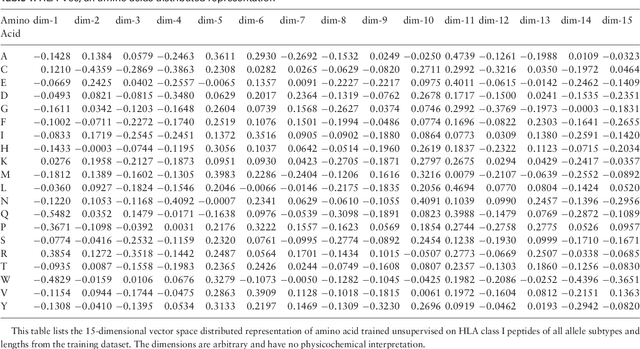
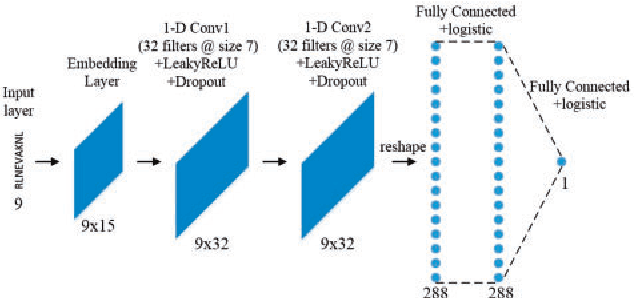
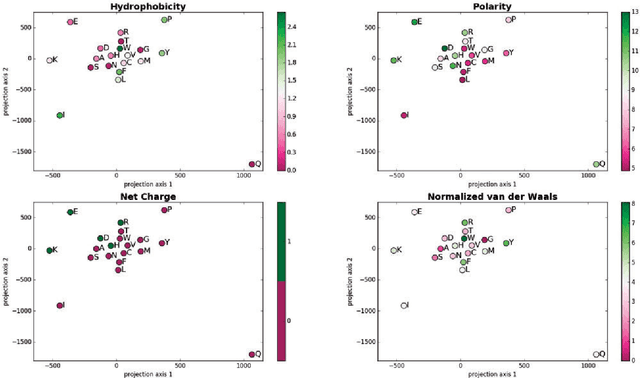
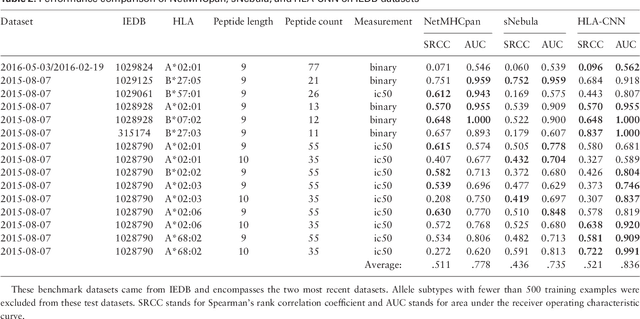
Abstract:Many biological processes are governed by protein-ligand interactions. One such example is the recognition of self and nonself cells by the immune system. This immune response process is regulated by the major histocompatibility complex (MHC) protein which is encoded by the human leukocyte antigen (HLA) complex. Understanding the binding potential between MHC and peptides can lead to the design of more potent, peptide-based vaccines and immunotherapies for infectious autoimmune diseases. We apply machine learning techniques from the natural language processing (NLP) domain to address the task of MHC-peptide binding prediction. More specifically, we introduce a new distributed representation of amino acids, name HLA-Vec, that can be used for a variety of downstream proteomic machine learning tasks. We then propose a deep convolutional neural network architecture, name HLA-CNN, for the task of HLA class I-peptide binding prediction. Experimental results show combining the new distributed representation with our HLA-CNN architecture achieves state-of-the-art results in the majority of the latest two Immune Epitope Database (IEDB) weekly automated benchmark datasets. We further apply our model to predict binding on the human genome and identify 15 genes with potential for self binding.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge