Yazhou Zhu
FAMNet: Frequency-aware Matching Network for Cross-domain Few-shot Medical Image Segmentation
Dec 12, 2024



Abstract:Existing few-shot medical image segmentation (FSMIS) models fail to address a practical issue in medical imaging: the domain shift caused by different imaging techniques, which limits the applicability to current FSMIS tasks. To overcome this limitation, we focus on the cross-domain few-shot medical image segmentation (CD-FSMIS) task, aiming to develop a generalized model capable of adapting to a broader range of medical image segmentation scenarios with limited labeled data from the novel target domain. Inspired by the characteristics of frequency domain similarity across different domains, we propose a Frequency-aware Matching Network (FAMNet), which includes two key components: a Frequency-aware Matching (FAM) module and a Multi-Spectral Fusion (MSF) module. The FAM module tackles two problems during the meta-learning phase: 1) intra-domain variance caused by the inherent support-query bias, due to the different appearances of organs and lesions, and 2) inter-domain variance caused by different medical imaging techniques. Additionally, we design an MSF module to integrate the different frequency features decoupled by the FAM module, and further mitigate the impact of inter-domain variance on the model's segmentation performance. Combining these two modules, our FAMNet surpasses existing FSMIS models and Cross-domain Few-shot Semantic Segmentation models on three cross-domain datasets, achieving state-of-the-art performance in the CD-FSMIS task.
RobustEMD: Domain Robust Matching for Cross-domain Few-shot Medical Image Segmentation
Oct 01, 2024



Abstract:Few-shot medical image segmentation (FSMIS) aims to perform the limited annotated data learning in the medical image analysis scope. Despite the progress has been achieved, current FSMIS models are all trained and deployed on the same data domain, as is not consistent with the clinical reality that medical imaging data is always across different data domains (e.g. imaging modalities, institutions and equipment sequences). How to enhance the FSMIS models to generalize well across the different specific medical imaging domains? In this paper, we focus on the matching mechanism of the few-shot semantic segmentation models and introduce an Earth Mover's Distance (EMD) calculation based domain robust matching mechanism for the cross-domain scenario. Specifically, we formulate the EMD transportation process between the foreground support-query features, the texture structure aware weights generation method, which proposes to perform the sobel based image gradient calculation over the nodes, is introduced in the EMD matching flow to restrain the domain relevant nodes. Besides, the point set level distance measurement metric is introduced to calculated the cost for the transportation from support set nodes to query set nodes. To evaluate the performance of our model, we conduct experiments on three scenarios (i.e., cross-modal, cross-sequence and cross-institution), which includes eight medical datasets and involves three body regions, and the results demonstrate that our model achieves the SoTA performance against the compared models.
Partition-A-Medical-Image: Extracting Multiple Representative Sub-regions for Few-shot Medical Image Segmentation
Sep 20, 2023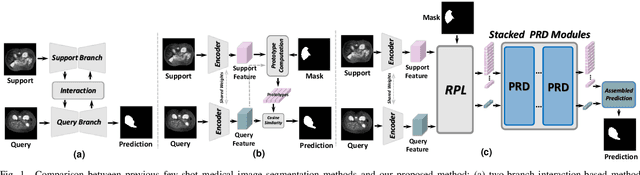
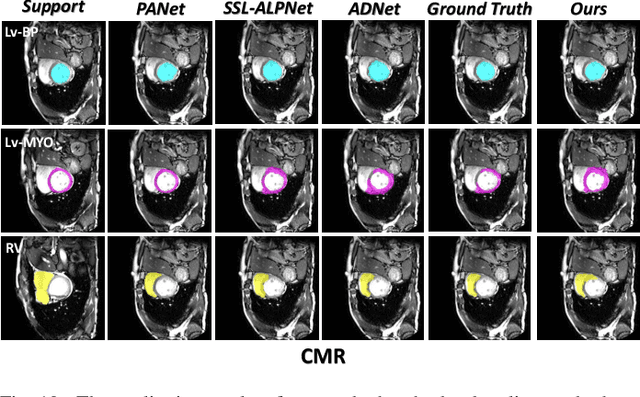
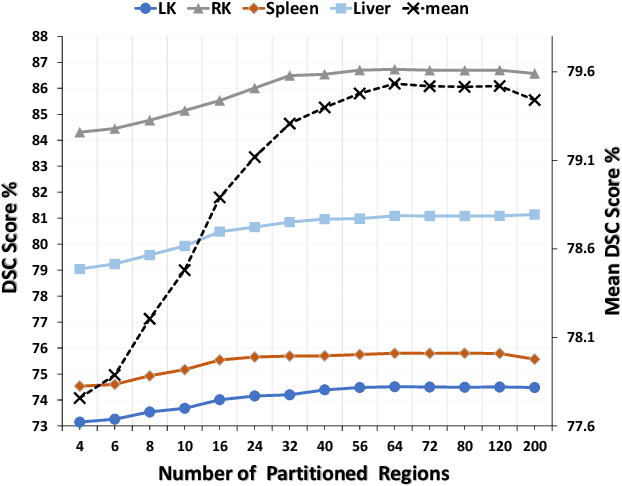
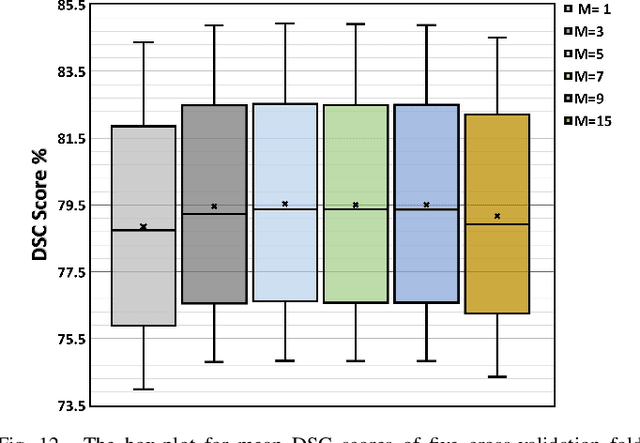
Abstract:Few-shot Medical Image Segmentation (FSMIS) is a more promising solution for medical image segmentation tasks where high-quality annotations are naturally scarce. However, current mainstream methods primarily focus on extracting holistic representations from support images with large intra-class variations in appearance and background, and encounter difficulties in adapting to query images. In this work, we present an approach to extract multiple representative sub-regions from a given support medical image, enabling fine-grained selection over the generated image regions. Specifically, the foreground of the support image is decomposed into distinct regions, which are subsequently used to derive region-level representations via a designed Regional Prototypical Learning (RPL) module. We then introduce a novel Prototypical Representation Debiasing (PRD) module based on a two-way elimination mechanism which suppresses the disturbance of regional representations by a self-support, Multi-direction Self-debiasing (MS) block, and a support-query, Interactive Debiasing (ID) block. Finally, an Assembled Prediction (AP) module is devised to balance and integrate predictions of multiple prototypical representations learned using stacked PRD modules. Results obtained through extensive experiments on three publicly accessible medical imaging datasets demonstrate consistent improvements over the leading FSMIS methods. The source code is available at https://github.com/YazhouZhu19/PAMI.
Few-Shot Medical Image Segmentation via a Region-enhanced Prototypical Transformer
Sep 09, 2023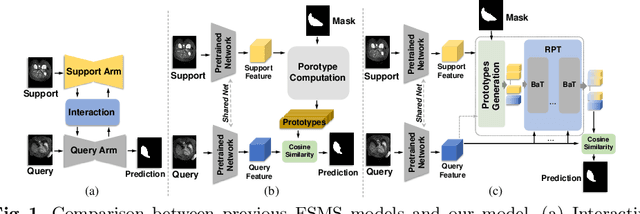
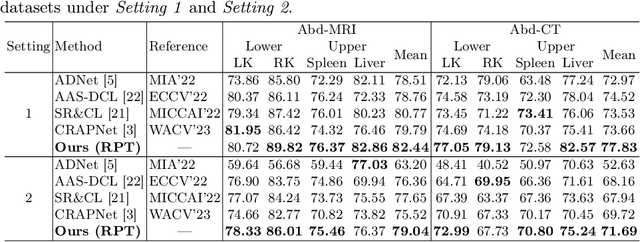

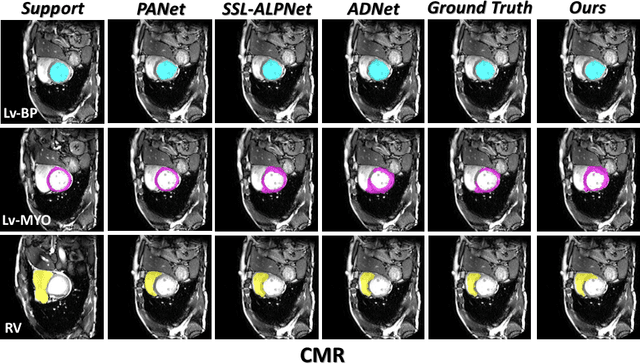
Abstract:Automated segmentation of large volumes of medical images is often plagued by the limited availability of fully annotated data and the diversity of organ surface properties resulting from the use of different acquisition protocols for different patients. In this paper, we introduce a more promising few-shot learning-based method named Region-enhanced Prototypical Transformer (RPT) to mitigate the effects of large intra-class diversity/bias. First, a subdivision strategy is introduced to produce a collection of regional prototypes from the foreground of the support prototype. Second, a self-selection mechanism is proposed to incorporate into the Bias-alleviated Transformer (BaT) block to suppress or remove interferences present in the query prototype and regional support prototypes. By stacking BaT blocks, the proposed RPT can iteratively optimize the generated regional prototypes and finally produce rectified and more accurate global prototypes for Few-Shot Medical Image Segmentation (FSMS). Extensive experiments are conducted on three publicly available medical image datasets, and the obtained results show consistent improvements compared to state-of-the-art FSMS methods. The source code is available at: https://github.com/YazhouZhu19/RPT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge