Yasmine Mustafa
Unlocking Neural Transparency: Jacobian Maps for Explainable AI in Alzheimer's Detection
Apr 04, 2025Abstract:Alzheimer's disease (AD) leads to progressive cognitive decline, making early detection crucial for effective intervention. While deep learning models have shown high accuracy in AD diagnosis, their lack of interpretability limits clinical trust and adoption. This paper introduces a novel pre-model approach leveraging Jacobian Maps (JMs) within a multi-modal framework to enhance explainability and trustworthiness in AD detection. By capturing localized brain volume changes, JMs establish meaningful correlations between model predictions and well-known neuroanatomical biomarkers of AD. We validate JMs through experiments comparing a 3D CNN trained on JMs versus on traditional preprocessed data, which demonstrates superior accuracy. We also employ 3D Grad-CAM analysis to provide both visual and quantitative insights, further showcasing improved interpretability and diagnostic reliability.
Efficient Brain Imaging Analysis for Alzheimer's and Dementia Detection Using Convolution-Derivative Operations
Nov 20, 2024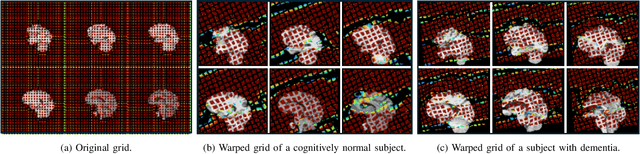
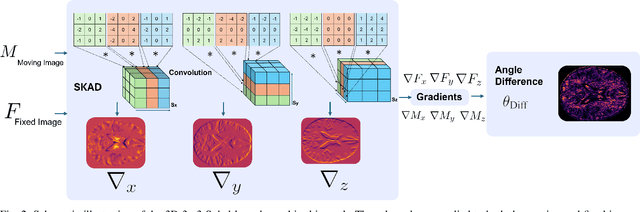
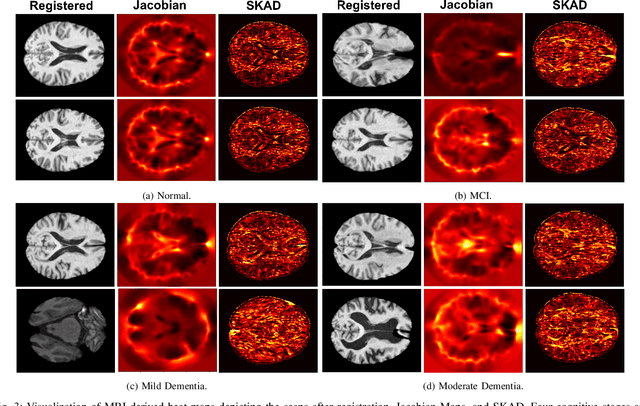
Abstract:Alzheimer's disease (AD) is characterized by progressive neurodegeneration and results in detrimental structural changes in human brains. Detecting these changes is crucial for early diagnosis and timely intervention of disease progression. Jacobian maps, derived from spatial normalization in voxel-based morphometry (VBM), have been instrumental in interpreting volume alterations associated with AD. However, the computational cost of generating Jacobian maps limits its clinical adoption. In this study, we explore alternative methods and propose Sobel kernel angle difference (SKAD) as a computationally efficient alternative. SKAD is a derivative operation that offers an optimized approach to quantifying volumetric alterations through localized analysis of the gradients. By efficiently extracting gradient amplitude changes at critical spatial regions, this derivative operation captures regional volume variations Evaluation of SKAD over various medical datasets demonstrates that it is 6.3x faster than Jacobian maps while still maintaining comparable accuracy. This makes it an efficient and competitive approach in neuroimaging research and clinical practice.
Unmasking Dementia Detection by Masking Input Gradients: A JSM Approach to Model Interpretability and Precision
Feb 25, 2024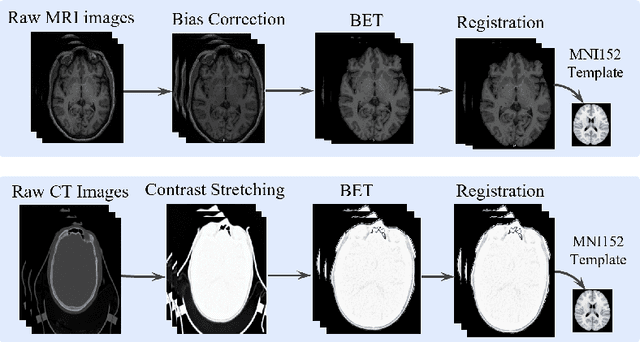
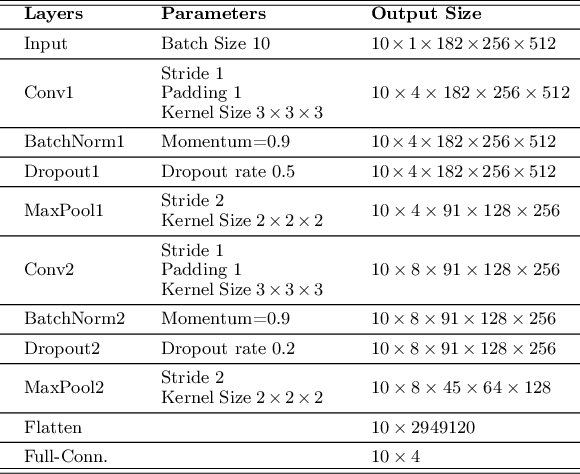
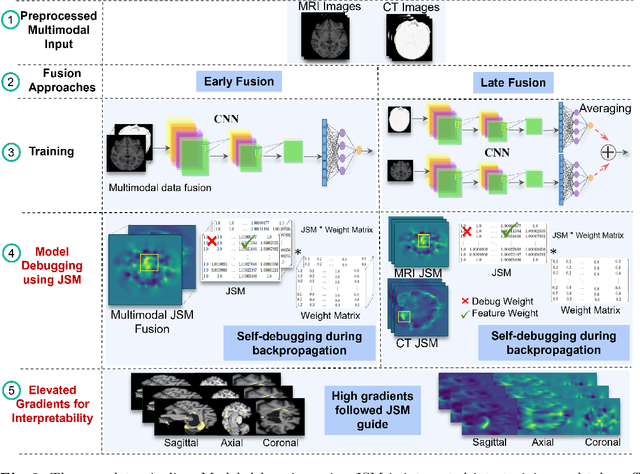
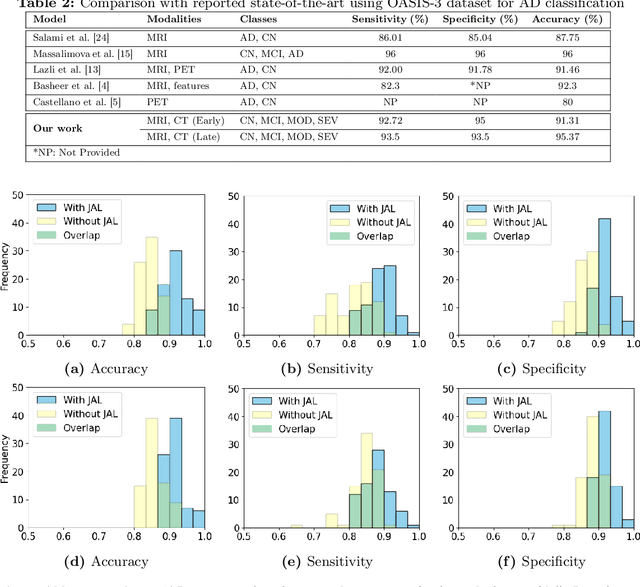
Abstract:The evolution of deep learning and artificial intelligence has significantly reshaped technological landscapes. However, their effective application in crucial sectors such as medicine demands more than just superior performance, but trustworthiness as well. While interpretability plays a pivotal role, existing explainable AI (XAI) approaches often do not reveal {\em Clever Hans} behavior where a model makes (ungeneralizable) correct predictions using spurious correlations or biases in data. Likewise, current post-hoc XAI methods are susceptible to generating unjustified counterfactual examples. In this paper, we approach XAI with an innovative {\em model debugging} methodology realized through Jacobian Saliency Map (JSM). To cast the problem into a concrete context, we employ Alzheimer's disease (AD) diagnosis as the use case, motivated by its significant impact on human lives and the formidable challenge in its early detection, stemming from the intricate nature of its progression. We introduce an interpretable, multimodal model for AD classification over its multi-stage progression, incorporating JSM as a modality-agnostic tool that provides insights into volumetric changes indicative of brain abnormalities. Our extensive evaluation including ablation study manifests the efficacy of using JSM for model debugging and interpretation, while significantly enhancing model accuracy as well.
Diagnosing Alzheimer's Disease using Early-Late Multimodal Data Fusion with Jacobian Maps
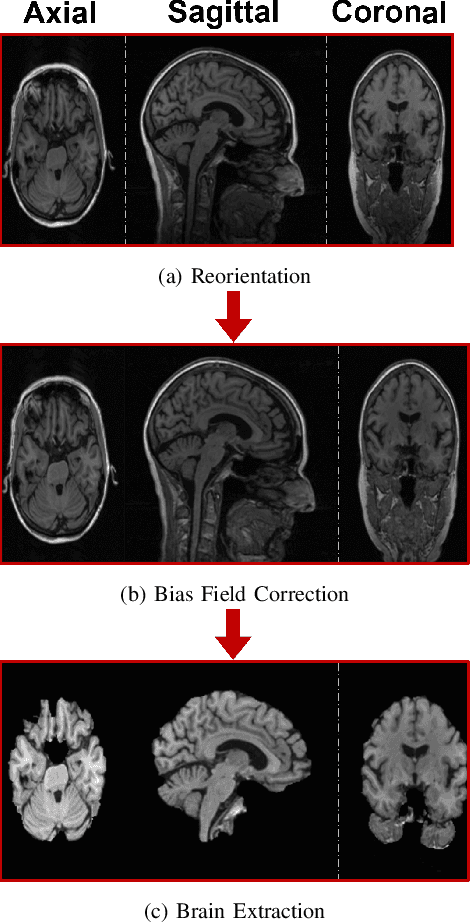
Oct 27, 2023Abstract:Alzheimer's disease (AD) is a prevalent and debilitating neurodegenerative disorder impacting a large aging population. Detecting AD in all its presymptomatic and symptomatic stages is crucial for early intervention and treatment. An active research direction is to explore machine learning methods that harness multimodal data fusion to outperform human inspection of medical scans. However, existing multimodal fusion models have limitations, including redundant computation, complex architecture, and simplistic handling of missing data. Moreover, the preprocessing pipelines of medical scans remain inadequately detailed and are seldom optimized for individual subjects. In this paper, we propose an efficient early-late fusion (ELF) approach, which leverages a convolutional neural network for automated feature extraction and random forests for their competitive performance on small datasets. Additionally, we introduce a robust preprocessing pipeline that adapts to the unique characteristics of individual subjects and makes use of whole brain images rather than slices or patches. Moreover, to tackle the challenge of detecting subtle changes in brain volume, we transform images into the Jacobian domain (JD) to enhance both accuracy and robustness in our classification. Using MRI and CT images from the OASIS-3 dataset, our experiments demonstrate the effectiveness of the ELF approach in classifying AD into four stages with an accuracy of 97.19%.
A Brain-Computer Interface Augmented Reality Framework with Auto-Adaptive SSVEP Recognition
Aug 11, 2023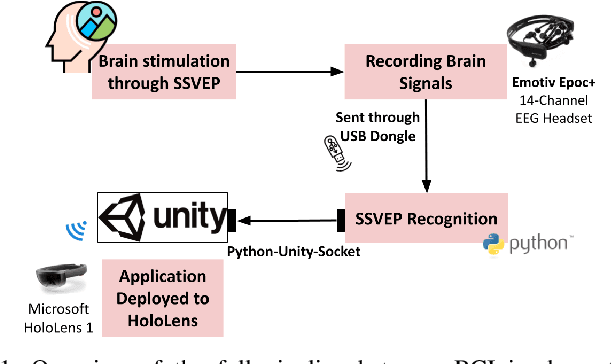
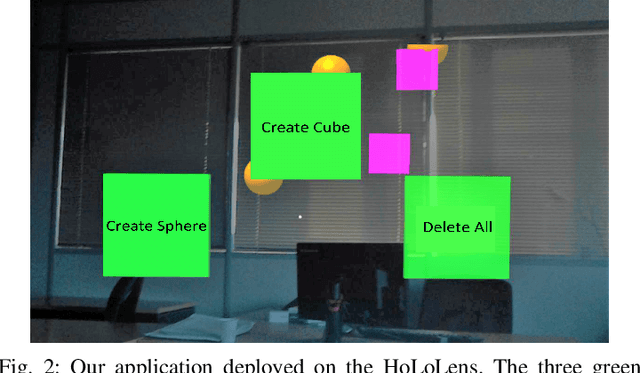
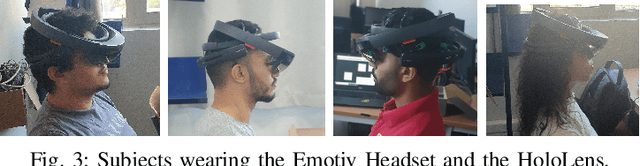
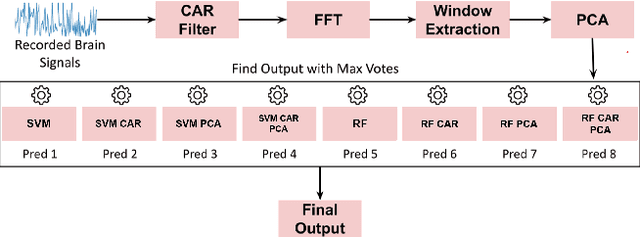
Abstract:Brain-Computer Interface (BCI) initially gained attention for developing applications that aid physically impaired individuals. Recently, the idea of integrating BCI with Augmented Reality (AR) emerged, which uses BCI not only to enhance the quality of life for individuals with disabilities but also to develop mainstream applications for healthy users. One commonly used BCI signal pattern is the Steady-state Visually-evoked Potential (SSVEP), which captures the brain's response to flickering visual stimuli. SSVEP-based BCI-AR applications enable users to express their needs/wants by simply looking at corresponding command options. However, individuals are different in brain signals and thus require per-subject SSVEP recognition. Moreover, muscle movements and eye blinks interfere with brain signals, and thus subjects are required to remain still during BCI experiments, which limits AR engagement. In this paper, we (1) propose a simple adaptive ensemble classification system that handles the inter-subject variability, (2) present a simple BCI-AR framework that supports the development of a wide range of SSVEP-based BCI-AR applications, and (3) evaluate the performance of our ensemble algorithm in an SSVEP-based BCI-AR application with head rotations which has demonstrated robustness to the movement interference. Our testing on multiple subjects achieved a mean accuracy of 80\% on a PC and 77\% using the HoloLens AR headset, both of which surpass previous studies that incorporate individual classifiers and head movements. In addition, our visual stimulation time is 5 seconds which is relatively short. The statistically significant results show that our ensemble classification approach outperforms individual classifiers in SSVEP-based BCIs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge