Yaping Zhu
Exploring Evolutionary Spectral Clustering for Temporal-Smoothed Clustered Cell-Free Networking
Dec 03, 2024Abstract:Clustered cell-free networking, which dynamically partitions the whole network into nonoverlapping subnetworks, has been recently proposed to mitigate the cell-edge problem in cellular networks. However, prior works only focused on optimizing clustered cell-free networking in static scenarios with fixed users. This could lead to a large number of handovers in the practical dynamic environment with moving users, seriously hindering the implementation of clustered cell-free networking in practice. This paper considers user mobility and aims to simultaneously maximize the sum rate and minimize the number of handovers. By transforming the multi-objective optimization problem into a time-varying graph partitioning problem and exploring evolutionary spectral clustering, a temporal-smoothed clustered cell-free networking algorithm is proposed, which is shown to be effective in smoothing network partitions over time and reducing handovers while maintaining similar sum rate.
PGD-UNet: A Position-Guided Deformable Network for Simultaneous Segmentation of Organs and Tumors
Jul 02, 2020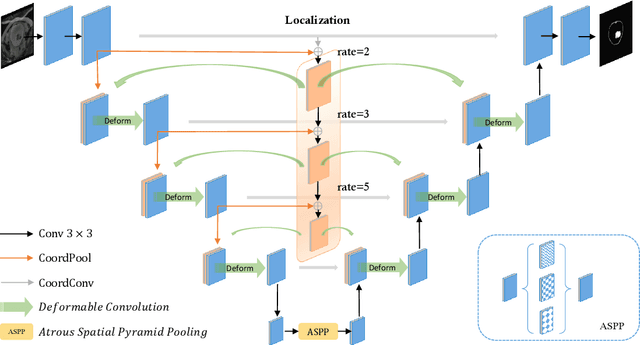
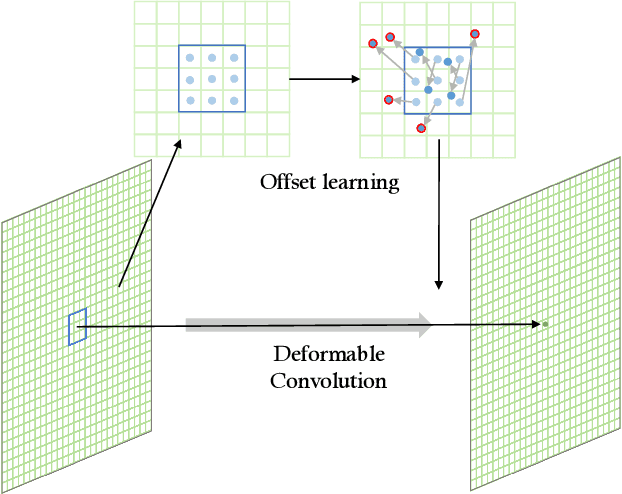
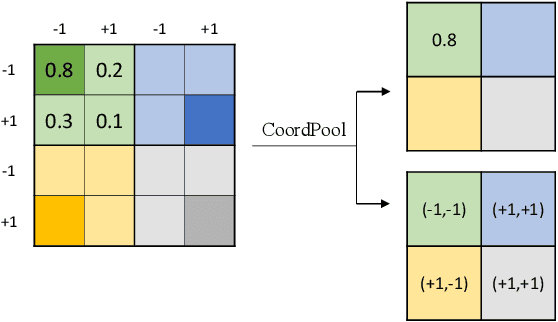
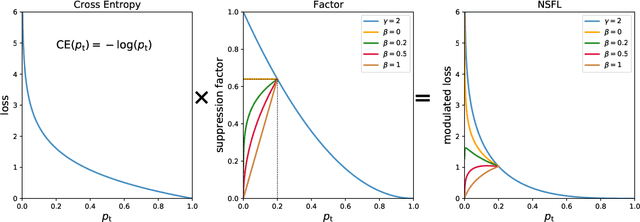
Abstract:Precise segmentation of organs and tumors plays a crucial role in clinical applications. It is a challenging task due to the irregular shapes and various sizes of organs and tumors as well as the significant class imbalance between the anatomy of interest (AOI) and the background region. In addition, in most situation tumors and normal organs often overlap in medical images, but current approaches fail to delineate both tumors and organs accurately. To tackle such challenges, we propose a position-guided deformable UNet, namely PGD-UNet, which exploits the spatial deformation capabilities of deformable convolution to deal with the geometric transformation of both organs and tumors. Position information is explicitly encoded into the network to enhance the capabilities of deformation. Meanwhile, we introduce a new pooling module to preserve position information lost in conventional max-pooling operation. Besides, due to unclear boundaries between different structures as well as the subjectivity of annotations, labels are not necessarily accurate for medical image segmentation tasks. It may cause the overfitting of the trained network due to label noise. To address this issue, we formulate a novel loss function to suppress the influence of potential label noise on the training process. Our method was evaluated on two challenging segmentation tasks and achieved very promising segmentation accuracy in both tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge