Wesley Yeung
Developing and validating multi-modal models for mortality prediction in COVID-19 patients: a multi-center retrospective study
Sep 01, 2021



Abstract:The unprecedented global crisis brought about by the COVID-19 pandemic has sparked numerous efforts to create predictive models for the detection and prognostication of SARS-CoV-2 infections with the goal of helping health systems allocate resources. Machine learning models, in particular, hold promise for their ability to leverage patient clinical information and medical images for prediction. However, most of the published COVID-19 prediction models thus far have little clinical utility due to methodological flaws and lack of appropriate validation. In this paper, we describe our methodology to develop and validate multi-modal models for COVID-19 mortality prediction using multi-center patient data. The models for COVID-19 mortality prediction were developed using retrospective data from Madrid, Spain (N=2547) and were externally validated in patient cohorts from a community hospital in New Jersey, USA (N=242) and an academic center in Seoul, Republic of Korea (N=336). The models we developed performed differently across various clinical settings, underscoring the need for a guided strategy when employing machine learning for clinical decision-making. We demonstrated that using features from both the structured electronic health records and chest X-ray imaging data resulted in better 30-day-mortality prediction performance across all three datasets (areas under the receiver operating characteristic curves: 0.85 (95% confidence interval: 0.83-0.87), 0.76 (0.70-0.82), and 0.95 (0.92-0.98)). We discuss the rationale for the decisions made at every step in developing the models and have made our code available to the research community. We employed the best machine learning practices for clinical model development. Our goal is to create a toolkit that would assist investigators and organizations in building multi-modal models for prediction, classification and/or optimization.
Prediction of Blood Lactate Values in Critically Ill Patients: A Retrospective Multi-center Cohort Study
Jul 07, 2021
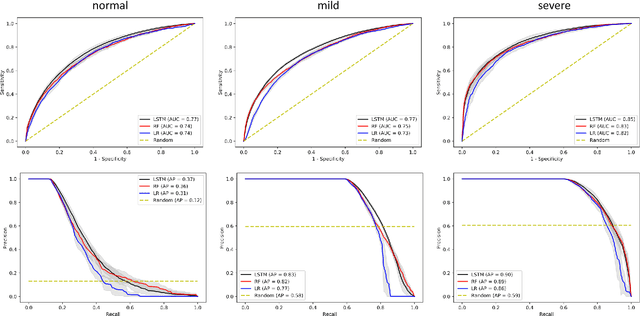
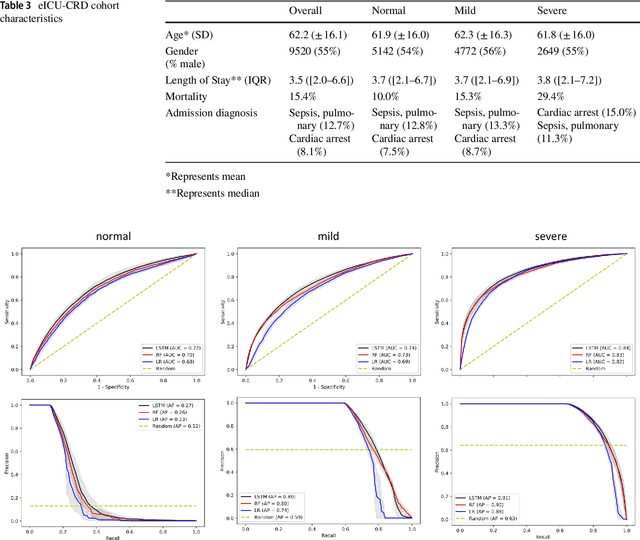
Abstract:Purpose. Elevations in initially obtained serum lactate levels are strong predictors of mortality in critically ill patients. Identifying patients whose serum lactate levels are more likely to increase can alert physicians to intensify care and guide them in the frequency of tending the blood test. We investigate whether machine learning models can predict subsequent serum lactate changes. Methods. We investigated serum lactate change prediction using the MIMIC-III and eICU-CRD datasets in internal as well as external validation of the eICU cohort on the MIMIC-III cohort. Three subgroups were defined based on the initial lactate levels: i) normal group (<2 mmol/L), ii) mild group (2-4 mmol/L), and iii) severe group (>4 mmol/L). Outcomes were defined based on increase or decrease of serum lactate levels between the groups. We also performed sensitivity analysis by defining the outcome as lactate change of >10% and furthermore investigated the influence of the time interval between subsequent lactate measurements on predictive performance. Results. The LSTM models were able to predict deterioration of serum lactate values of MIMIC-III patients with an AUC of 0.77 (95% CI 0.762-0.771) for the normal group, 0.77 (95% CI 0.768-0.772) for the mild group, and 0.85 (95% CI 0.840-0.851) for the severe group, with a slightly lower performance in the external validation. Conclusion. The LSTM demonstrated good discrimination of patients who had deterioration in serum lactate levels. Clinical studies are needed to evaluate whether utilization of a clinical decision support tool based on these results could positively impact decision-making and patient outcomes.
* 15 pages, 6 Appendices
An Emergency Medical Services Clinical Audit System driven by Named Entity Recognition from Deep Learning
Jul 07, 2020



Abstract:Clinical performance audits are routinely performed in Emergency Medical Services (EMS) to ensure adherence to treatment protocols, to identify individual areas of weakness for remediation, and to discover systemic deficiencies to guide the development of the training syllabus. At present, these audits are performed by manual chart review which is time-consuming and laborious. In this paper, we present an automatic audit system based on both the structured and unstructured ambulance case records and clinical notes with a deep neural network-based named entities recognition model. The dataset used in this study contained 58,898 unlabelled ambulance incidents encountered by the Singapore Civil Defence Force from 1st April 2019 to 30th June 2019. A weakly-supervised training approach was adopted to label the sentences. Later on, we trained three different models to perform the NER task. All three models achieve F1 scores of around 0.981 under entity type matching evaluation and around 0.976 under strict evaluation, while the BiLSTM-CRF model is 1~2 orders of magnitude lighter and faster than our BERT-based models. Overall, our approach yielded a named entity recognition model that could reliably identify clinical entities from unstructured paramedic free-text reports. Our proposed system may improve the efficiency of clinical performance audits and can also help with EMS database research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge