Viorel Mihalef
Graph convolutional regression of cardiac depolarization from sparse endocardial maps
Sep 28, 2020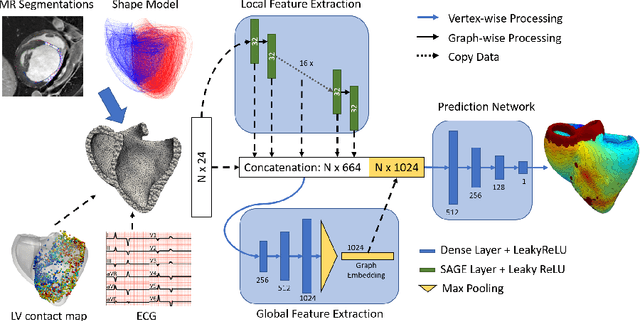

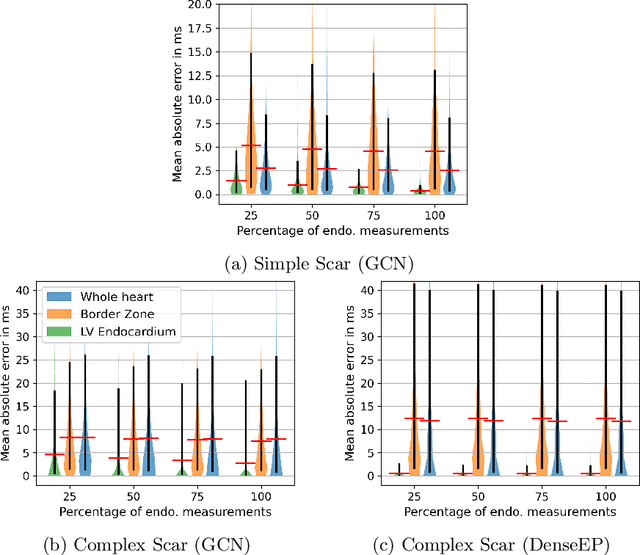
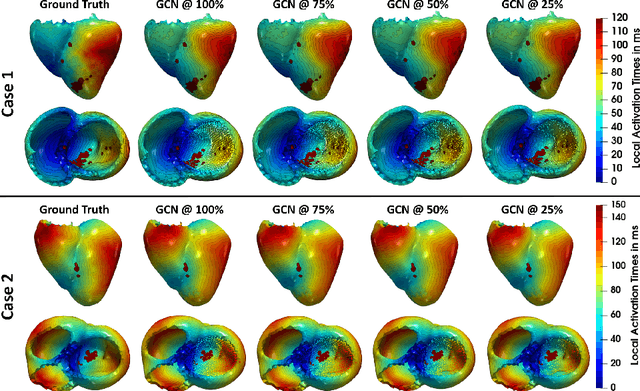
Abstract:Electroanatomic mapping as routinely acquired in ablation therapy of ventricular tachycardia is the gold standard method to identify the arrhythmogenic substrate. To reduce the acquisition time and still provide maps with high spatial resolution, we propose a novel deep learning method based on graph convolutional neural networks to estimate the depolarization time in the myocardium, given sparse catheter data on the left ventricular endocardium, ECG, and magnetic resonance images. The training set consists of data produced by a computational model of cardiac electrophysiology on a large cohort of synthetically generated geometries of ischemic hearts. The predicted depolarization pattern has good agreement with activation times computed by the cardiac electrophysiology model in a validation set of five swine heart geometries with complex scar and border zone morphologies. The mean absolute error hereby measures 8 ms on the entire myocardium when providing 50\% of the endocardial ground truth in over 500 computed depolarization patterns. Furthermore, when considering a complete animal data set with high density electroanatomic mapping data as reference, the neural network can accurately reproduce the endocardial depolarization pattern, even when a small percentage of measurements are provided as input features (mean absolute error of 7 ms with 50\% of input samples). The results show that the proposed method, trained on synthetically generated data, may generalize to real data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge