Vikram Singh
Indian Institute of Technology - Madras
Solving the Best Subset Selection Problem via Suboptimal Algorithms
Mar 31, 2025



Abstract:Best subset selection in linear regression is well known to be nonconvex and computationally challenging to solve, as the number of possible subsets grows rapidly with increasing dimensionality of the problem. As a result, finding the global optimal solution via an exact optimization method for a problem with dimensions of 1000s may take an impractical amount of CPU time. This suggests the importance of finding suboptimal procedures that can provide good approximate solutions using much less computational effort than exact methods. In this work, we introduce a new procedure and compare it with other popular suboptimal algorithms to solve the best subset selection problem. Extensive computational experiments using synthetic and real data have been performed. The results provide insights into the performance of these methods in different data settings. The new procedure is observed to be a competitive suboptimal algorithm for solving the best subset selection problem for high-dimensional data.
Higher order organizational features can distinguish protein interaction networks of disease classes: a case study of neoplasms and neurological diseases
Dec 26, 2022



Abstract:Neoplasms (NPs) and neurological diseases and disorders (NDDs) are amongst the major classes of diseases underlying deaths of a disproportionate number of people worldwide. To determine if there exist some distinctive features in the local wiring patterns of protein interactions emerging at the onset of a disease belonging to either of these two classes, we examined 112 and 175 protein interaction networks belonging to NPs and NDDs, respectively. Orbit usage profiles (OUPs) for each of these networks were enumerated by investigating the networks' local topology. 56 non-redundant OUPs (nrOUPs) were derived and used as network features for classification between these two disease classes. Four machine learning classifiers, namely, k-nearest neighbour (KNN), support vector machine (SVM), deep neural network (DNN), random forest (RF) were trained on these data. DNN obtained the greatest average AUPRC (0.988) among these classifiers. DNNs developed on node2vec and the proposed nrOUPs embeddings were compared using 5-fold cross validation on the basis of average values of the six of performance measures, viz., AUPRC, Accuracy, Sensitivity, Specificity, Precision and MCC. It was found that nrOUPs based classifier performed better in all of these six performance measures.
Characterizing the organizational diversity of protein interaction networks across three domains of life
Mar 02, 2022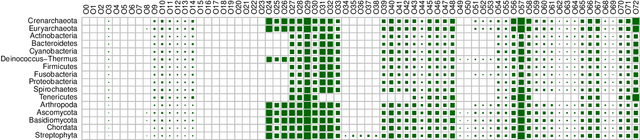
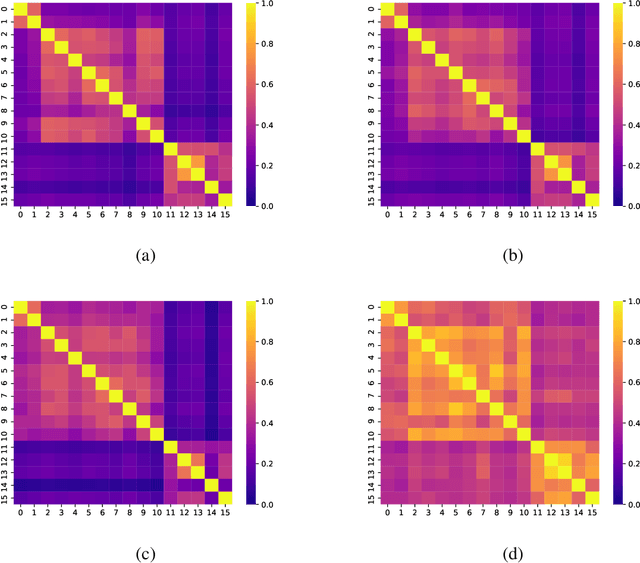
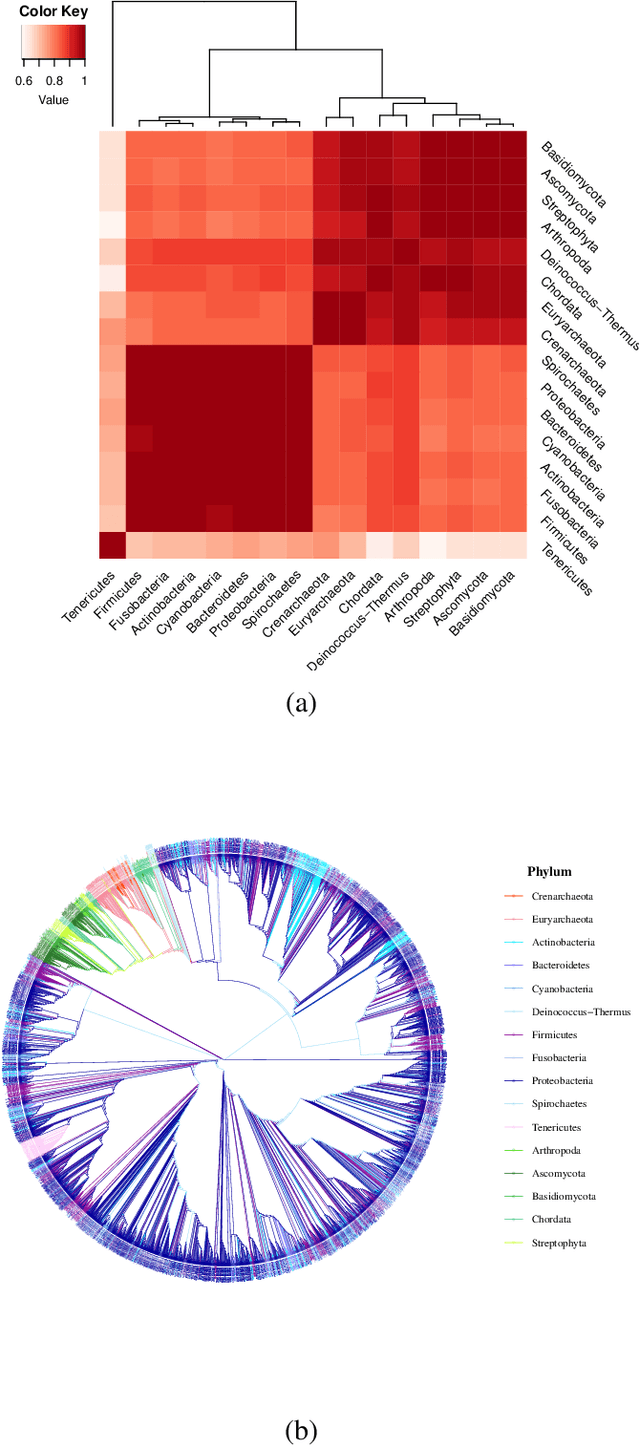
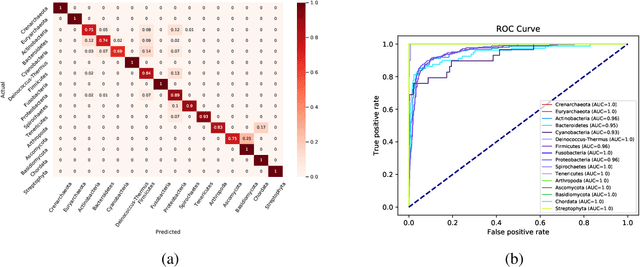
Abstract:Networks exist everywhere in nature from the physical, chemical, biological or social worlds to the designed spheres. To explore, if there exists some higher-order organization that can be exploited to distinguish different types of networks, we study 4,738 protein interaction networks (PINs) belonging to 16 phyla encompassing all the three domains of life. Our method utilizes positional information of a network's nodes by appropriately normalizing the frequency of automorphic orbits appearing in the induced graphlets of sizes 2-5. There are some evolutionary constraints imposed on the network's topology which shape its local architecture as well as its behavior. According to these rules (features), each type of network occupies its respective position within a common network space. A deep neural network was trained on differentially expressed orbits resulting in a prediction accuracy of 85%. Our results indicate that nature has, probably, allocated a specific band of design space to various superfamilies of PINs.
WDN: A Wide and Deep Network to Divide-and-Conquer Image Super-resolution
Oct 07, 2020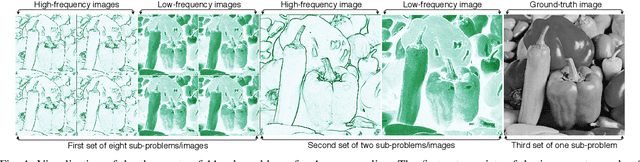
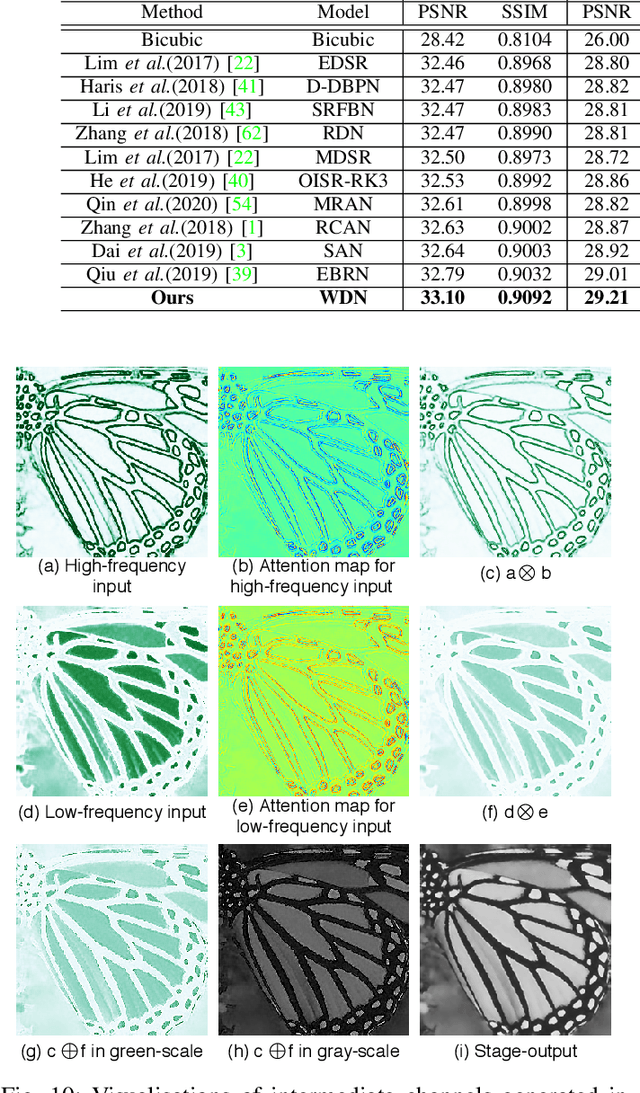
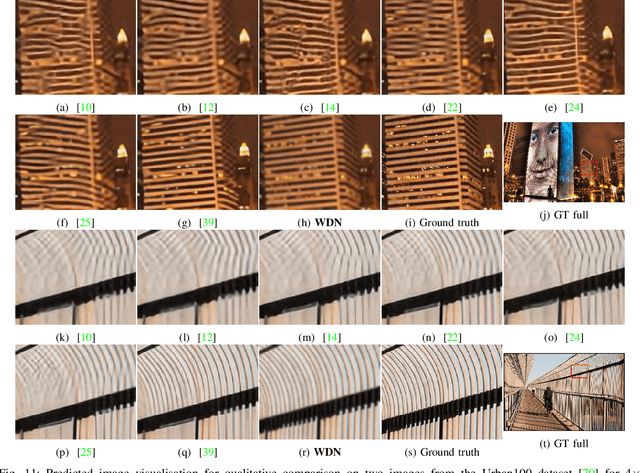
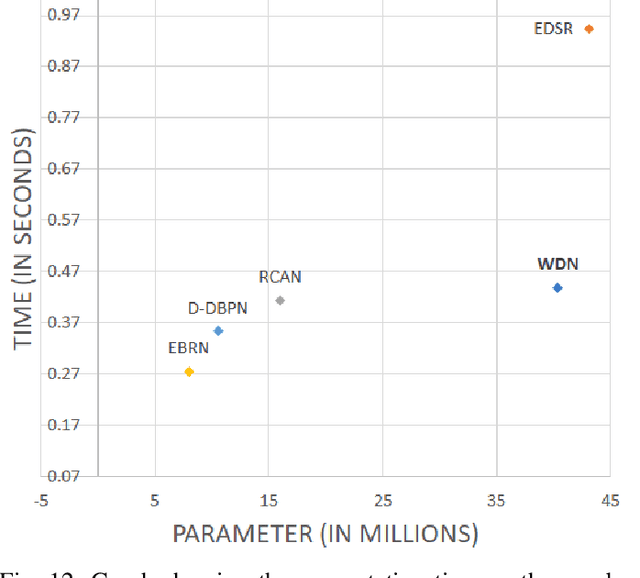
Abstract:Divide and conquer is an established algorithm design paradigm that has proven itself to solve a variety of problems efficiently. However, it is yet to be fully explored in solving problems with a neural network, particularly the problem of image super-resolution. In this work, we propose an approach to divide the problem of image super-resolution into multiple sub-problems and then solve/conquer them with the help of a neural network. Unlike a typical deep neural network, we design an alternate network architecture that is much wider (along with being deeper) than existing networks and is specially designed to implement the divide-and-conquer design paradigm with a neural network. Additionally, a technique to calibrate the intensities of feature map pixels is being introduced. Extensive experimentation on five datasets reveals that our approach towards the problem and the proposed architecture generate better and sharper results than current state-of-the-art methods.
Integrating User's Domain Knowledge with Association Rule Mining
Apr 20, 2010



Abstract:This paper presents a variation of Apriori algorithm that includes the role of domain expert to guide and speed up the overall knowledge discovery task. Usually, the user is interested in finding relationships between certain attributes instead of the whole dataset. Moreover, he can help the mining algorithm to select the target database which in turn takes less time to find the desired association rules. Variants of the standard Apriori and Interactive Apriori algorithms have been run on artificial datasets. The results show that incorporating user's preference in selection of target attribute helps to search the association rules efficiently both in terms of space and time.
* International Journal of Computer Science Issues online at http://ijcsi.org/articles/Integrating-Users-Domain-Knowledge-with-Association-Rule-Mining.php
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge