Characterizing the organizational diversity of protein interaction networks across three domains of life
Paper and Code
Mar 02, 2022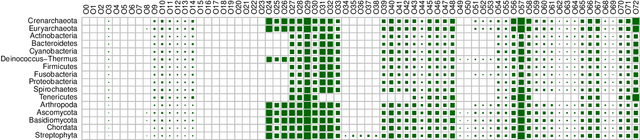
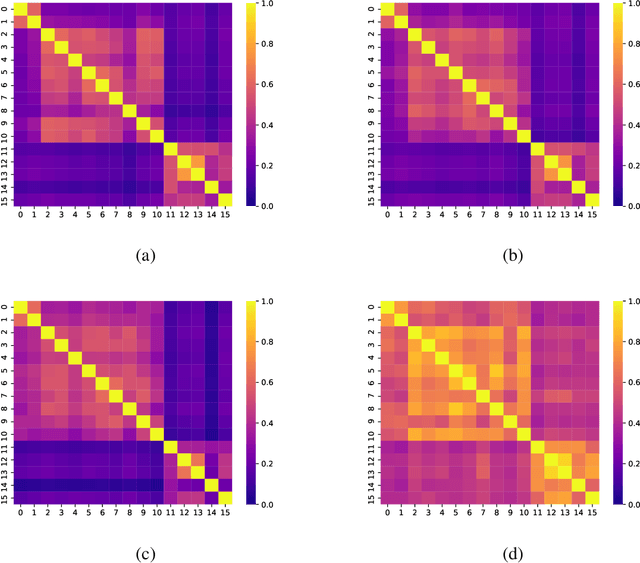
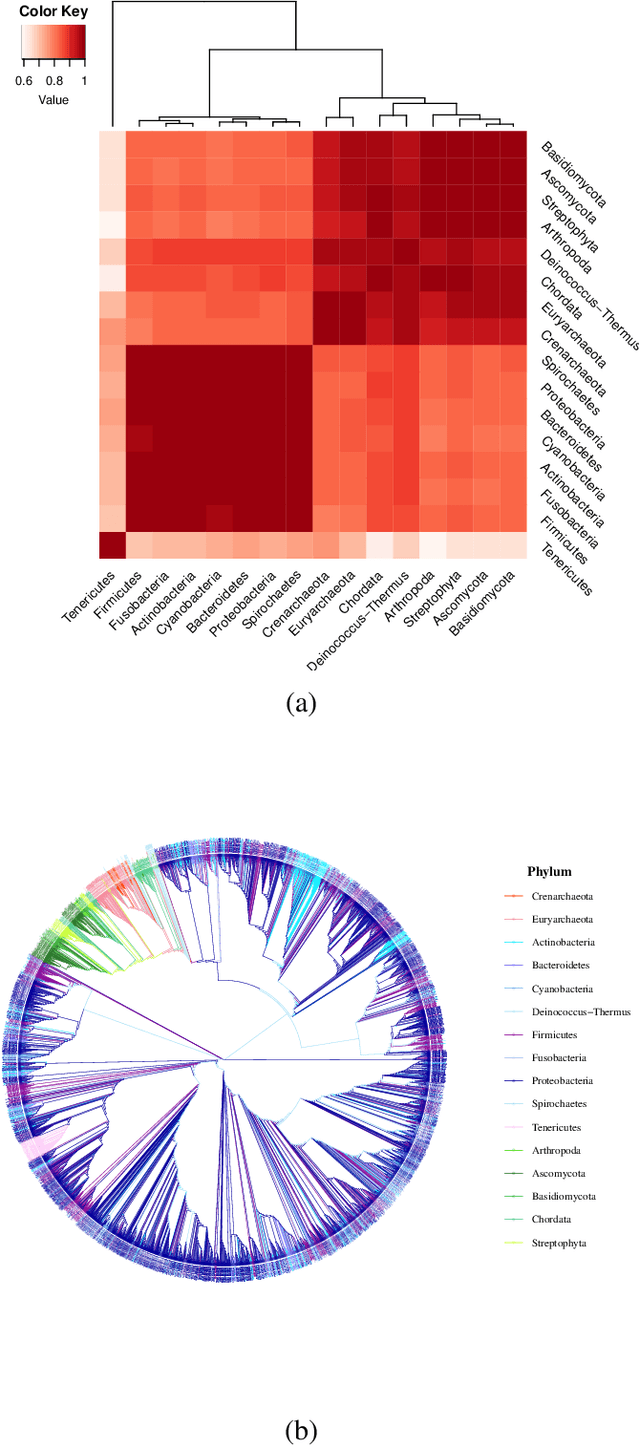
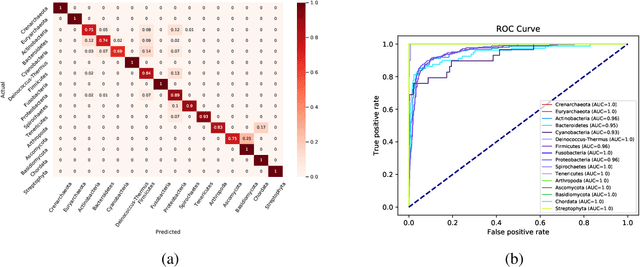
Networks exist everywhere in nature from the physical, chemical, biological or social worlds to the designed spheres. To explore, if there exists some higher-order organization that can be exploited to distinguish different types of networks, we study 4,738 protein interaction networks (PINs) belonging to 16 phyla encompassing all the three domains of life. Our method utilizes positional information of a network's nodes by appropriately normalizing the frequency of automorphic orbits appearing in the induced graphlets of sizes 2-5. There are some evolutionary constraints imposed on the network's topology which shape its local architecture as well as its behavior. According to these rules (features), each type of network occupies its respective position within a common network space. A deep neural network was trained on differentially expressed orbits resulting in a prediction accuracy of 85%. Our results indicate that nature has, probably, allocated a specific band of design space to various superfamilies of PINs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge