Vaibhav Sahai
Inferring Clinically Relevant Molecular Subtypes of Pancreatic Cancer from Routine Histopathology Using Deep Learning
Jan 06, 2026Abstract:Molecular subtyping of PDAC into basal-like and classical has established prognostic and predictive value. However, its use in clinical practice is limited by cost, turnaround time, and tissue requirements, thereby restricting its application in the management of PDAC. We introduce PanSubNet, an interpretable deep learning framework that predicts therapy-relevant molecular subtypes directly from standard H&E-stained WSIs. PanSubNet was developed using data from 1,055 patients across two multi-institutional cohorts (PANCAN, n=846; TCGA, n=209) with paired histology and RNA-seq data. Ground-truth labels were derived using the validated Moffitt 50-gene signature refined by GATA6 expression. The model employs dual-scale architecture that fuses cellular-level morphology with tissue-level architecture, leveraging attention mechanisms for multi-scale representation learning and transparent feature attribution. On internal validation within PANCAN using five-fold cross-validation, PanSubNet achieved mean AUC of 88.5% with balanced sensitivity and specificity. External validation on the independent TCGA cohort without fine-tuning demonstrated robust generalizability (AUC 84.0%). PanSubNet preserved and, in metastatic disease, strengthened prognostic stratification compared to RNA-seq based labels. Prediction uncertainty linked to intermediate transcriptional states, not classification noise. Model predictions are aligned with established transcriptomic programs, differentiation markers, and DNA damage repair signatures. By enabling rapid, cost-effective molecular stratification from routine H&E-stained slides, PanSubNet offers a clinically deployable and interpretable tool for genetic subtyping. We are gathering data from two institutions to validate and assess real-world performance, supporting integration into digital pathology workflows and advancing precision oncology for PDAC.
Democratizing LLMs: An Exploration of Cost-Performance Trade-offs in Self-Refined Open-Source Models
Oct 22, 2023

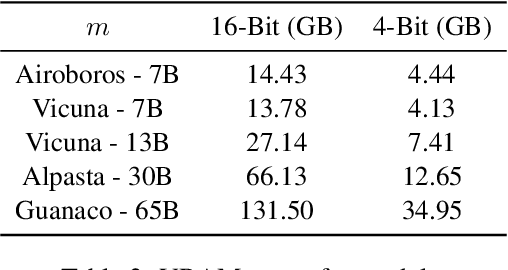
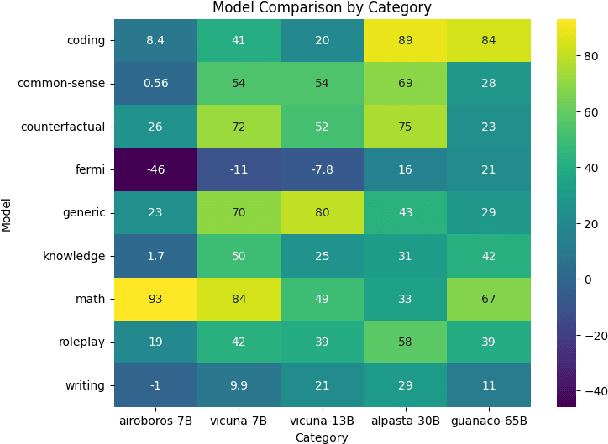
Abstract:The dominance of proprietary LLMs has led to restricted access and raised information privacy concerns. High-performing open-source alternatives are crucial for information-sensitive and high-volume applications but often lag behind in performance. To address this gap, we propose (1) A untargeted variant of iterative self-critique and self-refinement devoid of external influence. (2) A novel ranking metric - Performance, Refinement, and Inference Cost Score (PeRFICS) - to find the optimal model for a given task considering refined performance and cost. Our experiments show that SoTA open source models of varying sizes from 7B - 65B, on average, improve 8.2% from their baseline performance. Strikingly, even models with extremely small memory footprints, such as Vicuna-7B, show a 11.74% improvement overall and up to a 25.39% improvement in high-creativity, open ended tasks on the Vicuna benchmark. Vicuna-13B takes it a step further and outperforms ChatGPT post-refinement. This work has profound implications for resource-constrained and information-sensitive environments seeking to leverage LLMs without incurring prohibitive costs, compromising on performance and privacy. The domain-agnostic self-refinement process coupled with our novel ranking metric facilitates informed decision-making in model selection, thereby reducing costs and democratizing access to high-performing language models, as evidenced by case studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge