Usma Niyaz
Understanding Calibration of Deep Neural Networks for Medical Image Classification
Sep 22, 2023Abstract:In the field of medical image analysis, achieving high accuracy is not enough; ensuring well-calibrated predictions is also crucial. Confidence scores of a deep neural network play a pivotal role in explainability by providing insights into the model's certainty, identifying cases that require attention, and establishing trust in its predictions. Consequently, the significance of a well-calibrated model becomes paramount in the medical imaging domain, where accurate and reliable predictions are of utmost importance. While there has been a significant effort towards training modern deep neural networks to achieve high accuracy on medical imaging tasks, model calibration and factors that affect it remain under-explored. To address this, we conducted a comprehensive empirical study that explores model performance and calibration under different training regimes. We considered fully supervised training, which is the prevailing approach in the community, as well as rotation-based self-supervised method with and without transfer learning, across various datasets and architecture sizes. Multiple calibration metrics were employed to gain a holistic understanding of model calibration. Our study reveals that factors such as weight distributions and the similarity of learned representations correlate with the calibration trends observed in the models. Notably, models trained using rotation-based self-supervised pretrained regime exhibit significantly better calibration while achieving comparable or even superior performance compared to fully supervised models across different medical imaging datasets. These findings shed light on the importance of model calibration in medical image analysis and highlight the benefits of incorporating self-supervised learning approach to improve both performance and calibration.
Leveraging Different Learning Styles for Improved Knowledge Distillation
Dec 06, 2022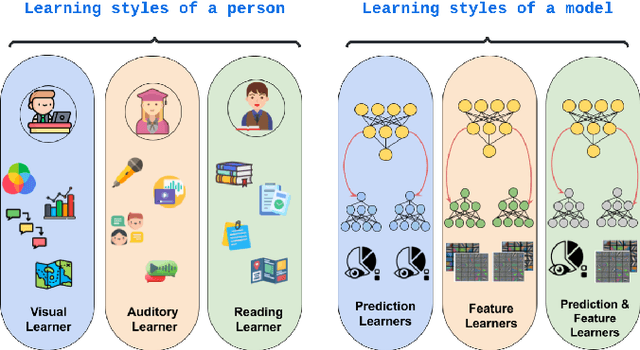
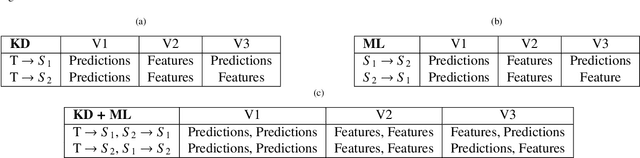
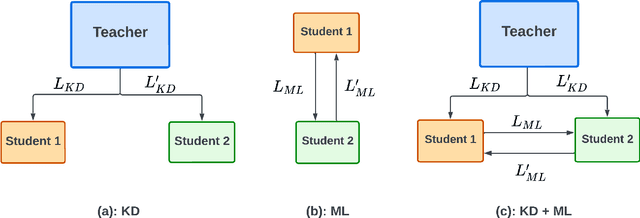
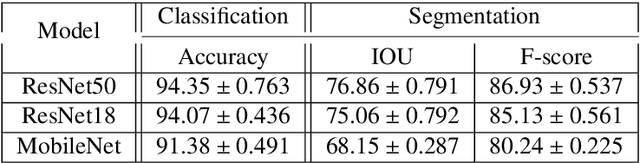
Abstract:Learning style refers to a type of training mechanism adopted by an individual to gain new knowledge. As suggested by the VARK model, humans have different learning preferences like visual, auditory, etc., for acquiring and effectively processing information. Inspired by this concept, our work explores the idea of mixed information sharing with model compression in the context of Knowledge Distillation (KD) and Mutual Learning (ML). Unlike conventional techniques that share the same type of knowledge with all networks, we propose to train individual networks with different forms of information to enhance the learning process. We formulate a combined KD and ML framework with one teacher and two student networks that share or exchange information in the form of predictions and feature maps. Our comprehensive experiments with benchmark classification and segmentation datasets demonstrate that with 15% compression, the ensemble performance of networks trained with diverse forms of knowledge outperforms the conventional techniques both quantitatively and qualitatively.
Augmenting Knowledge Distillation With Peer-To-Peer Mutual Learning For Model Compression
Oct 22, 2021



Abstract:Knowledge distillation (KD) is an effective model compression technique where a compact student network is taught to mimic the behavior of a complex and highly trained teacher network. In contrast, Mutual Learning (ML) provides an alternative strategy where multiple simple student networks benefit from sharing knowledge, even in the absence of a powerful but static teacher network. Motivated by these findings, we propose a single-teacher, multi-student framework that leverages both KD and ML to achieve better performance. Furthermore, an online distillation strategy is utilized to train the teacher and students simultaneously. To evaluate the performance of the proposed approach, extensive experiments were conducted using three different versions of teacher-student networks on benchmark biomedical classification (MSI vs. MSS) and object detection (Polyp Detection) tasks. Ensemble of student networks trained in the proposed manner achieved better results than the ensemble of students trained using KD or ML individually, establishing the benefit of augmenting knowledge transfer from teacher to students with peer-to-peer learning between students.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge