Uli Niemann
Training and Validating a Treatment Recommender with Partial Verification Evidence
Jun 10, 2024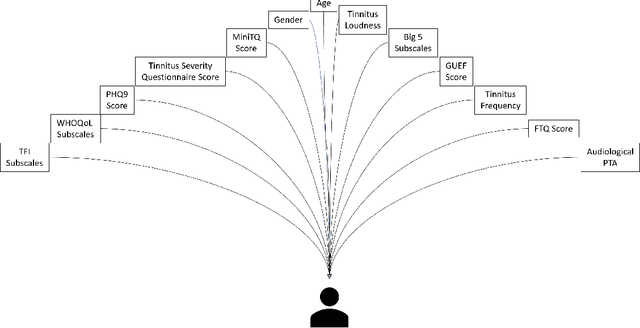

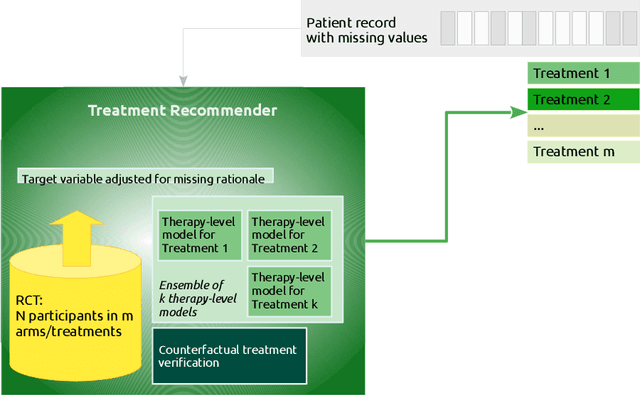

Abstract:Current clinical decision support systems (DSS) are trained and validated on observational data from the target clinic. This is problematic for treatments validated in a randomized clinical trial (RCT), but not yet introduced in any clinic. In this work, we report on a method for training and validating the DSS using the RCT data. The key challenges we address are of missingness -- missing rationale for treatment assignment (the assignment is at random), and missing verification evidence, since the effectiveness of a treatment for a patient can only be verified (ground truth) for treatments what were actually assigned to a patient. We use data from a multi-armed RCT that investigated the effectiveness of single- and combination- treatments for 240+ tinnitus patients recruited and treated in 5 clinical centers. To deal with the 'missing rationale' challenge, we re-model the target variable (outcome) in order to suppress the effect of the randomly-assigned treatment, and control on the effect of treatment in general. Our methods are also robust to missing values in features and with a small number of patients per RCT arm. We deal with 'missing verification evidence' by using counterfactual treatment verification, which compares the effectiveness of the DSS recommendations to the effectiveness of the RCT assignments when they are aligned v/s not aligned. We demonstrate that our approach leverages the RCT data for learning and verification, by showing that the DSS suggests treatments that improve the outcome. The results are limited through the small number of patients per treatment; while our ensemble is designed to mitigate this effect, the predictive performance of the methods is affected by the smallness of the data. We provide a basis for the establishment of decision supporting routines on treatments that have been tested in RCTs but have not yet been deployed clinically.
A cost-based multi-layer network approach for the discovery of patient phenotypes
Sep 20, 2022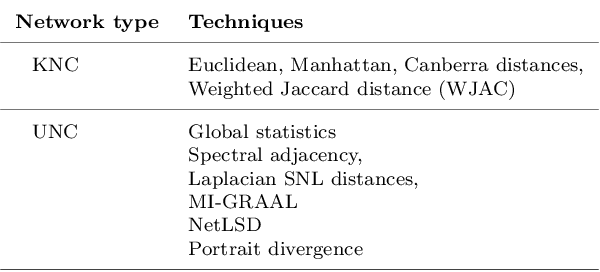
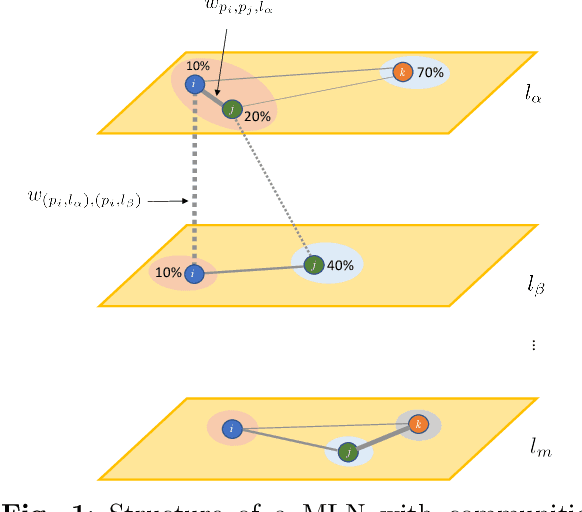
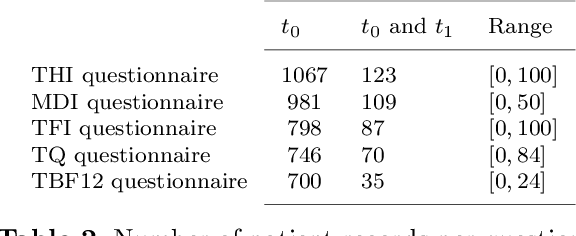
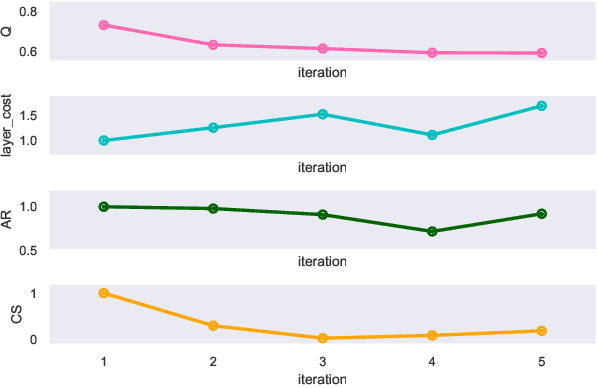
Abstract:Clinical records frequently include assessments of the characteristics of patients, which may include the completion of various questionnaires. These questionnaires provide a variety of perspectives on a patient's current state of well-being. Not only is it critical to capture the heterogeneity given by these perspectives, but there is also a growing demand for developing cost-effective technologies for clinical phenotyping. Filling out many questionnaires may be a strain for the patients and therefore costly. In this work, we propose COBALT -- a cost-based layer selector model for detecting phenotypes using a community detection approach. Our goal is to minimize the number of features used to build these phenotypes while preserving its quality. We test our model using questionnaire data from chronic tinnitus patients and represent the data in a multi-layer network structure. The model is then evaluated by predicting post-treatment data using baseline features (age, gender, and pre-treatment data) as well as the identified phenotypes as a feature. For some post-treatment variables, predictors using phenotypes from COBALT as features outperformed those using phenotypes detected by traditional clustering methods. Moreover, using phenotype data to predict post-treatment data proved beneficial in comparison with predictors that were solely trained with baseline features.
Cardiac Cohort Classification based on Morphologic and Hemodynamic Parameters extracted from 4D PC-MRI Data
Oct 12, 2020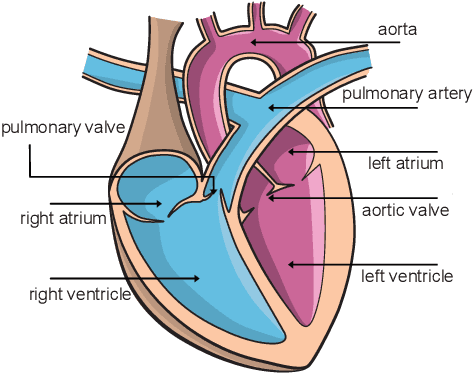

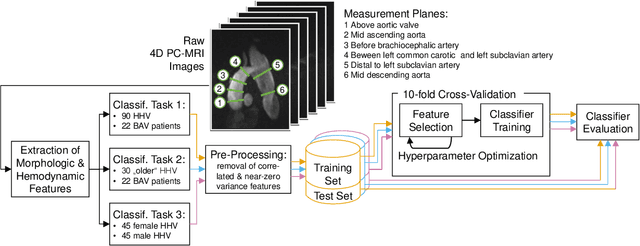

Abstract:An accurate assessment of the cardiovascular system and prediction of cardiovascular diseases (CVDs) are crucial. Measured cardiac blood flow data provide insights about patient-specific hemodynamics, where many specialized techniques have been developed for the visual exploration of such data sets to better understand the influence of morphological and hemodynamic conditions on CVDs. However, there is a lack of machine learning approaches techniques that allow a feature-based classification of heart-healthy people and patients with CVDs.In this work, we investigate the potential of morphological and hemodynamic characteristics, extracted from measured blood flow data in the aorta, for the classification of heart-healthy volunteers and patients with bicuspid aortic valve (BAV). Furthermore, we research if there are characteristic features to classify male and female as well as younger and older heart-healthy volunteers. We propose a data analysis pipeline for the classification of the cardiac status, encompassing feature selection, model training and hyperparameter tuning. In our experiments, we use several feature selection methods and classification algorithms to train separate models for the healthy subgroups and BAV patients. We report on classification performance and investigate the predictive power of morphological and hemodynamic features with regard to the classification oft he defined groups. Finally, we identify the key features for the best models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge