Trevor McKeown
Medical Physics Program, Duke University
A Vessel Bifurcation Landmark Pair Dataset for Abdominal CT Deformable Image Registration (DIR) Validation
Jan 15, 2025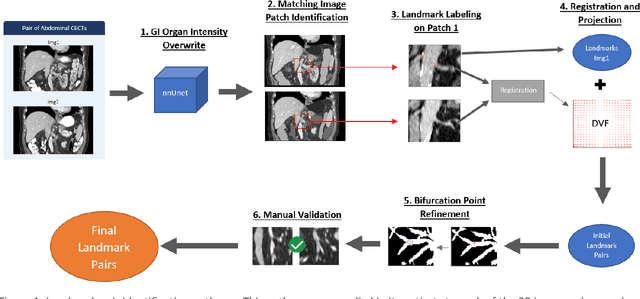
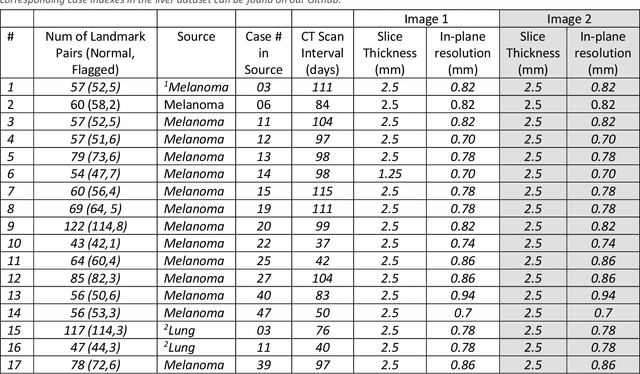
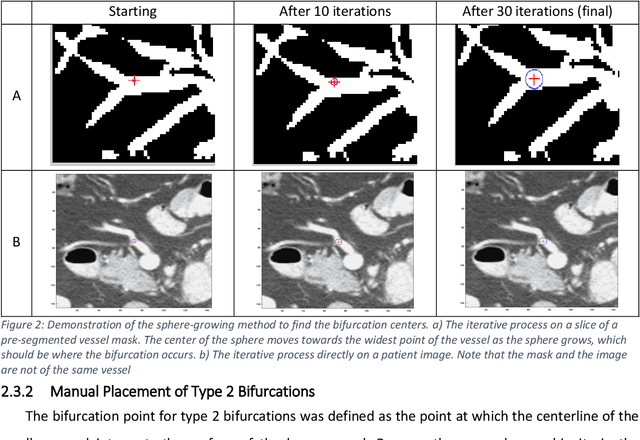
Abstract:Deformable image registration (DIR) is an enabling technology in many diagnostic and therapeutic tasks. Despite this, DIR algorithms have limited clinical use, largely due to a lack of benchmark datasets for quality assurance during development. To support future algorithm development, here we introduce our first-of-its-kind abdominal CT DIR benchmark dataset, comprising large numbers of highly accurate landmark pairs on matching blood vessel bifurcations. Abdominal CT image pairs of 30 patients were acquired from several public repositories as well as the authors' institution with IRB approval. The two CTs of each pair were originally acquired for the same patient on different days. An image processing workflow was developed and applied to each image pair: 1) Abdominal organs were segmented with a deep learning model, and image intensity within organ masks was overwritten. 2) Matching image patches were manually identified between two CTs of each image pair 3) Vessel bifurcation landmarks were labeled on one image of each image patch pair. 4) Image patches were deformably registered, and landmarks were projected onto the second image. 5) Landmark pair locations were refined manually or with an automated process. This workflow resulted in 1895 total landmark pairs, or 63 per case on average. Estimates of the landmark pair accuracy using digital phantoms were 0.7+/-1.2mm. The data is published in Zenodo at https://doi.org/10.5281/zenodo.14362785. Instructions for use can be found at https://github.com/deshanyang/Abdominal-DIR-QA. This dataset is a first-of-its-kind for abdominal DIR validation. The number, accuracy, and distribution of landmark pairs will allow for robust validation of DIR algorithms with precision beyond what is currently available.
Small metal artifact detection and inpainting in cardiac CT images
Sep 25, 2024

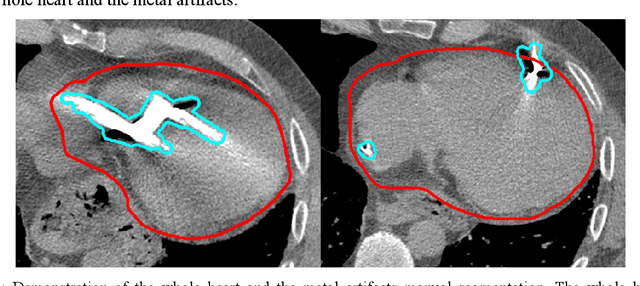

Abstract:Background: Quantification of cardiac motion on pre-treatment CT imaging for stereotactic arrhythmia radiotherapy patients is difficult due to the presence of image artifacts caused by metal leads of implantable cardioverter-defibrillators (ICDs). New methods are needed to accurately reduce the metal artifacts in already reconstructed CTs to recover the otherwise lost anatomical information. Purpose: To develop a methodology to automatically detect metal artifacts in cardiac CT scans and inpaint the affected volume with anatomically consistent structures and values. Methods: ECG-gated 4DCT scans of 12 patients who underwent cardiac radiation therapy for treating ventricular tachycardia were collected. The metal artifacts in the images were manually contoured. A 2D U-Net deep learning (DL) model was developed to segment the metal artifacts. A dataset of synthetic CTs was prepared by adding metal artifacts from the patient images to artifact-free CTs. A 3D image inpainting DL model was trained to refill the metal artifact portion in the synthetic images with realistic values. The inpainting model was evaluated by analyzing the automated segmentation results of the four heart chambers on the synthetic dataset. Additionally, the raw cardiac patient cases were qualitatively inspected. Results: The artifact detection model produced a Dice score of 0.958 +- 0.008. The inpainting model was able to recreate images with a structural similarity index of 0.988 +- 0.012. With the chamber segmentations improved surface Dice scores from 0.684 +- 0.247 to 0.964 +- 0.067 and the Hausdorff distance reduced from 3.4 +- 3.9 mm to 0.7 +- 0.7 mm. The inpainting model's use on cardiac patient CTs was visually inspected and the artifact-inpainted images were visually plausible. Conclusion: We successfully developed two deep models to detect and inpaint metal artifacts in cardiac CT images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge