Tien Huynh
In-Pocket 3D Graphs Enhance Ligand-Target Compatibility in Generative Small-Molecule Creation
Apr 05, 2022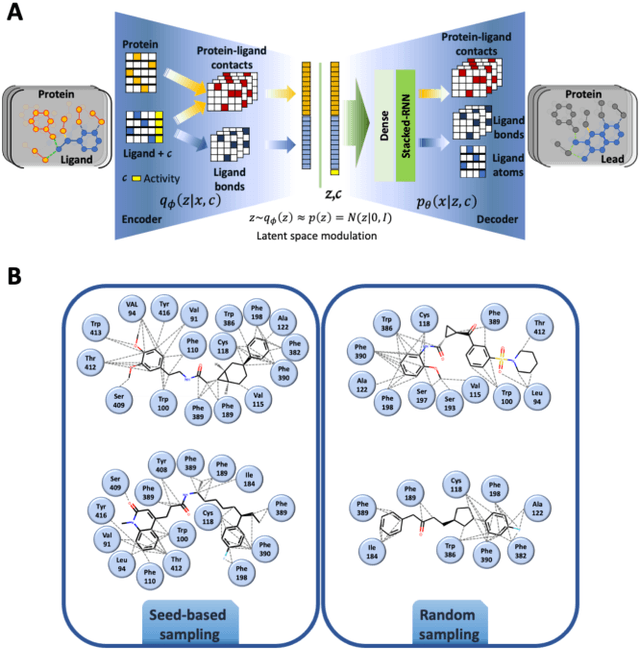
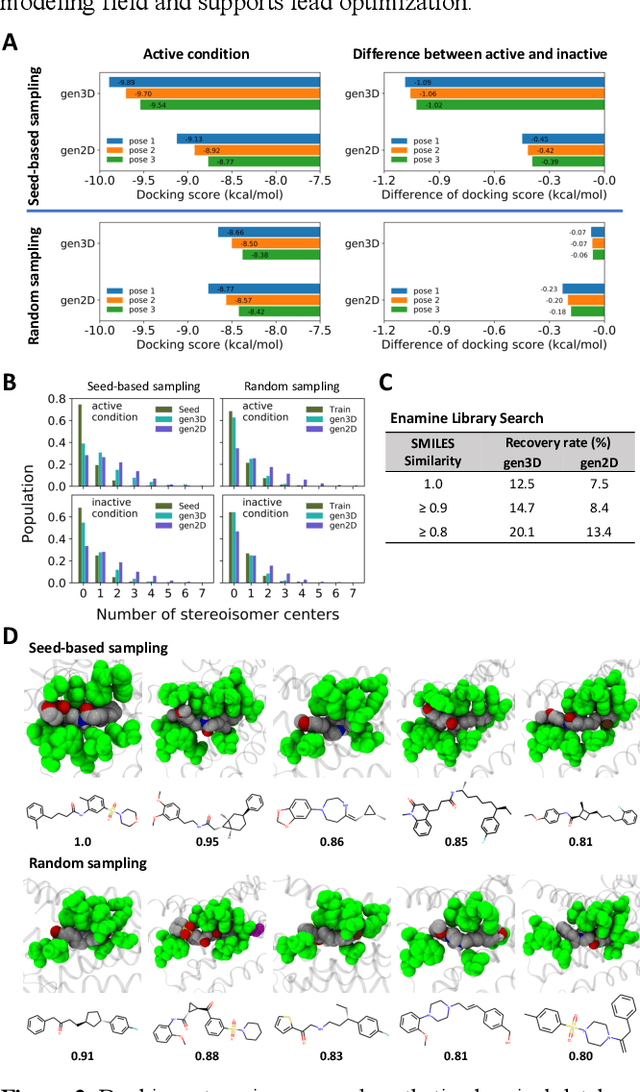

Abstract:Proteins in complex with small molecule ligands represent the core of structure-based drug discovery. However, three-dimensional representations are absent from most deep-learning-based generative models. We here present a graph-based generative modeling technology that encodes explicit 3D protein-ligand contacts within a relational graph architecture. The models combine a conditional variational autoencoder that allows for activity-specific molecule generation with putative contact generation that provides predictions of molecular interactions within the target binding pocket. We show that molecules generated with our 3D procedure are more compatible with the binding pocket of the dopamine D2 receptor than those produced by a comparable ligand-based 2D generative method, as measured by docking scores, expected stereochemistry, and recoverability in commercial chemical databases. Predicted protein-ligand contacts were found among highest-ranked docking poses with a high recovery rate. This work shows how the structural context of a protein target can be used to enhance molecule generation.
Combining docking pose rank and structure with deep learning improves protein-ligand binding mode prediction
Oct 07, 2019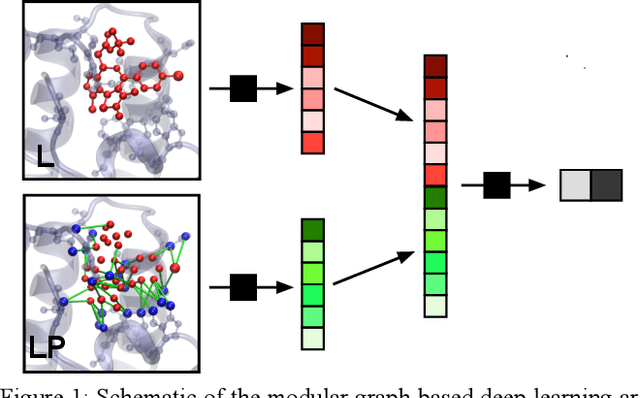
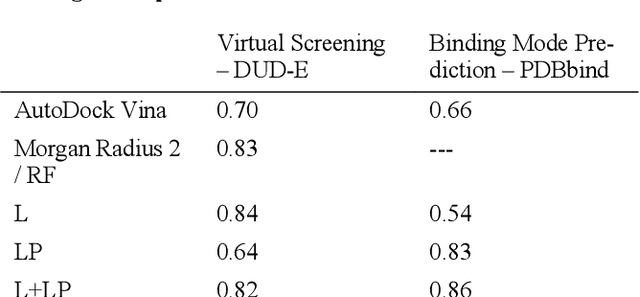
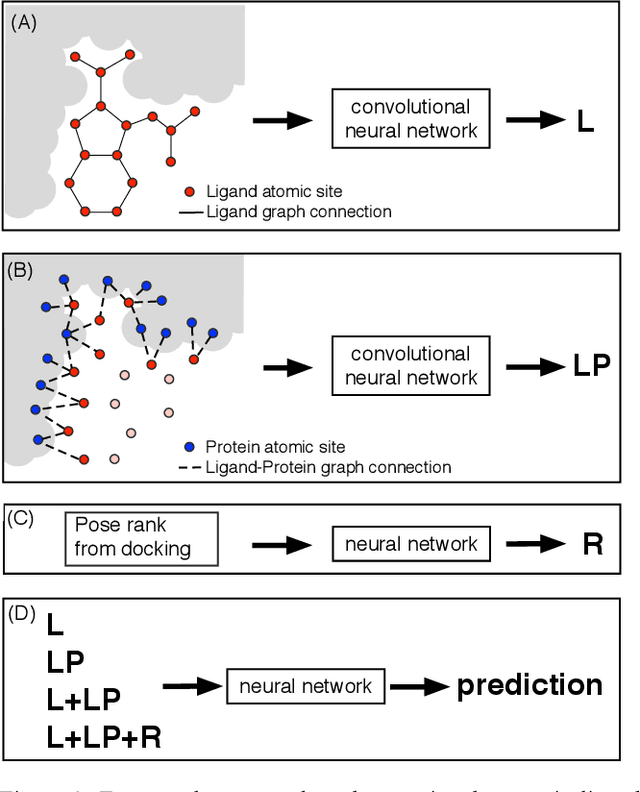
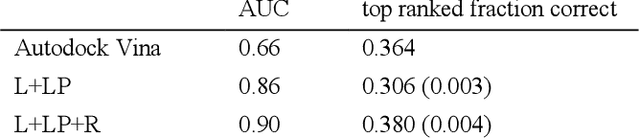
Abstract:We present a simple, modular graph-based convolutional neural network that takes structural information from protein-ligand complexes as input to generate models for activity and binding mode prediction. Complex structures are generated by a standard docking procedure and fed into a dual-graph architecture that includes separate sub-networks for the ligand bonded topology and the ligand-protein contact map. This network division allows contributions from ligand identity to be distinguished from effects of protein-ligand interactions on classification. We show, in agreement with recent literature, that dataset bias drives many of the promising results on virtual screening that have previously been reported. However, we also show that our neural network is capable of learning from protein structural information when, as in the case of binding mode prediction, an unbiased dataset is constructed. We develop a deep learning model for binding mode prediction that uses docking ranking as input in combination with docking structures. This strategy mirrors past consensus models and outperforms the baseline docking program in a variety of tests, including on cross-docking datasets that mimic real-world docking use cases. Furthermore, the magnitudes of network predictions serve as reliable measures of model confidence
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge