Tibo Duran
Uncovering the Mechanism of Hepatotoxiciy of PFAS Targeting L-FABP Using GCN and Computational Modeling
Sep 16, 2024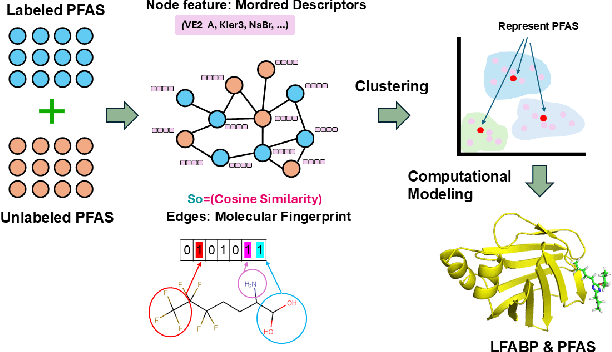
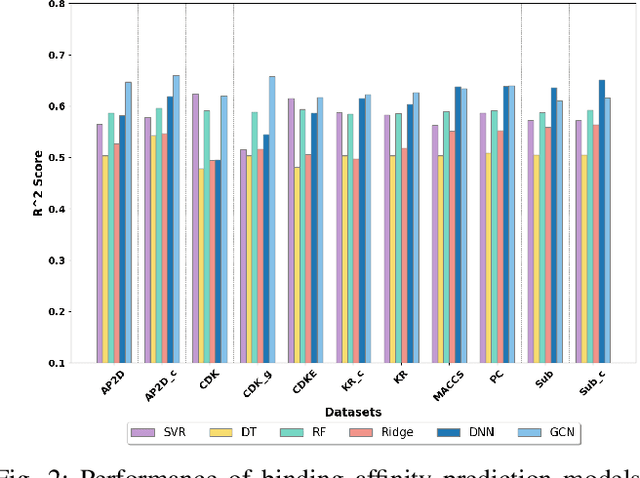
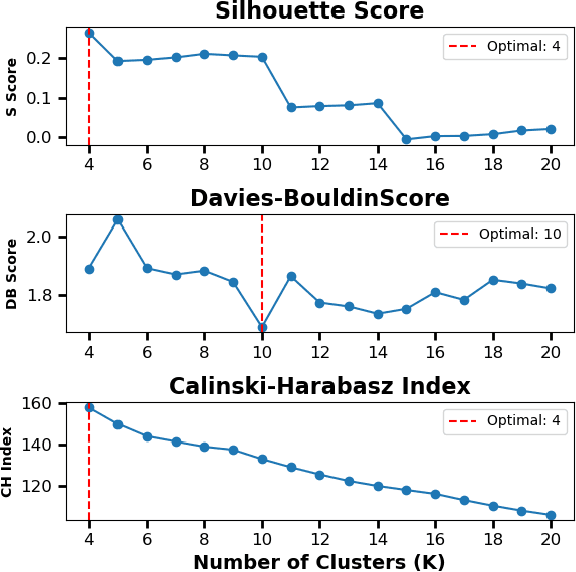
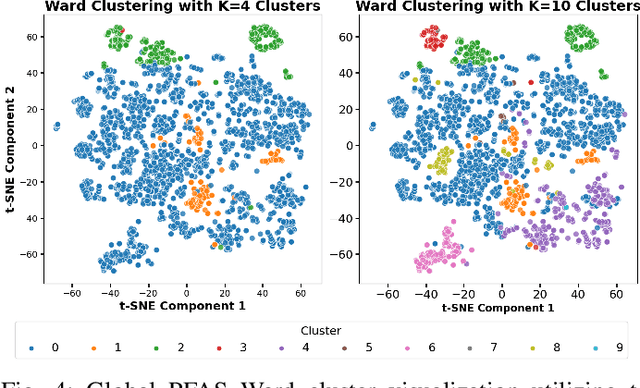
Abstract:Per- and polyfluoroalkyl substances (PFAS) are persistent environmental pollutants with known toxicity and bioaccumulation issues. Their widespread industrial use and resistance to degradation have led to global environmental contamination and significant health concerns. While a minority of PFAS have been extensively studied, the toxicity of many PFAS remains poorly understood due to limited direct toxicological data. This study advances the predictive modeling of PFAS toxicity by combining semi-supervised graph convolutional networks (GCNs) with molecular descriptors and fingerprints. We propose a novel approach to enhance the prediction of PFAS binding affinities by isolating molecular fingerprints to construct graphs where then descriptors are set as the node features. This approach specifically captures the structural, physicochemical, and topological features of PFAS without overfitting due to an abundance of features. Unsupervised clustering then identifies representative compounds for detailed binding studies. Our results provide a more accurate ability to estimate PFAS hepatotoxicity to provide guidance in chemical discovery of new PFAS and the development of new safety regulations.
Exploring Latent Space for Generating Peptide Analogs Using Protein Language Models
Aug 15, 2024
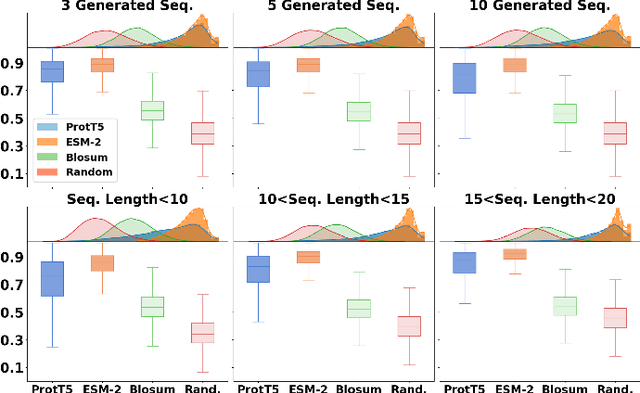


Abstract:Generating peptides with desired properties is crucial for drug discovery and biotechnology. Traditional sequence-based and structure-based methods often require extensive datasets, which limits their effectiveness. In this study, we proposed a novel method that utilized autoencoder shaped models to explore the protein embedding space, and generate novel peptide analogs by leveraging protein language models. The proposed method requires only a single sequence of interest, avoiding the need for large datasets. Our results show significant improvements over baseline models in similarity indicators of peptide structures, descriptors and bioactivities. The proposed method validated through Molecular Dynamics simulations on TIGIT inhibitors, demonstrates that our method produces peptide analogs with similar yet distinct properties, highlighting its potential to enhance peptide screening processes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge