Terumasa Tokunaga
LEA-Net: Layer-wise External Attention Network for Efficient Color Anomaly Detection
Sep 12, 2021



Abstract:The utilization of prior knowledge about anomalies is an essential issue for anomaly detections. Recently, the visual attention mechanism has become a promising way to improve the performance of CNNs for some computer vision tasks. In this paper, we propose a novel model called Layer-wise External Attention Network (LEA-Net) for efficient image anomaly detection. The core idea relies on the integration of unsupervised and supervised anomaly detectors via the visual attention mechanism. Our strategy is as follows: (i) Prior knowledge about anomalies is represented as the anomaly map generated by unsupervised learning of normal instances, (ii) The anomaly map is translated to an attention map by the external network, (iii) The attention map is then incorporated into intermediate layers of the anomaly detection network. Notably, this layer-wise external attention can be applied to any CNN model in an end-to-end training manner. For a pilot study, we validate LEA-Net on color anomaly detection tasks. Through extensive experiments on PlantVillage, MVTec AD, and Cloud datasets, we demonstrate that the proposed layer-wise visual attention mechanism consistently boosts anomaly detection performances of an existing CNN model, even on imbalanced datasets. Moreover, we show that our attention mechanism successfully boosts the performance of several CNN models.
Image-based plant disease diagonasis with unsupervised anomaly detection based on reconstructability of colors
Dec 22, 2020



Abstract:This paper proposes an unsupervised anomaly detection technique for image-based plant disease diagnosis. A construction of large and openly available data set on labeled images of healthy and diseased crop plants has led to growing interest in computer vision techniques for automatic plant disease diagnosis. Although supervised image classifiers based on deep learning could be a powerful tool to identify plant diseases, they require huge amount of data sets that have been labeled as healthy and diseased. While, data mining techniques called "anomaly detection" include unsupervised approaches that not require rare samples for training classifiers. The proposed method in this study focuses on the reconstructability of colors on plant images. We expect that a deep encoder decoder network trained for reconstructing colors of healthy plant images certainly fails to reconstruct colors of symptomatic regions. The main contributions of this work are as follows: (i) we propose a new image-based plant disease detection framework utilizing a conditional adversarial network called pix2pix, (ii) we introduce a new anomaly score based on CIEDE2000 color difference. Through experiments using PlantVillage dataset, we demonstrate that our method is superior to an existing anomaly detector called AnoGAN for identifying diseased crop images in terms of accuracy, interpretability and computational efficiency.
SPF-CellTracker: Tracking multiple cells with strongly-correlated moves using a spatial particle filter
May 27, 2016
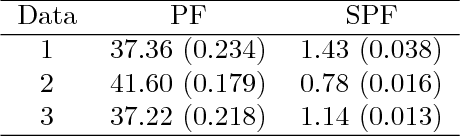
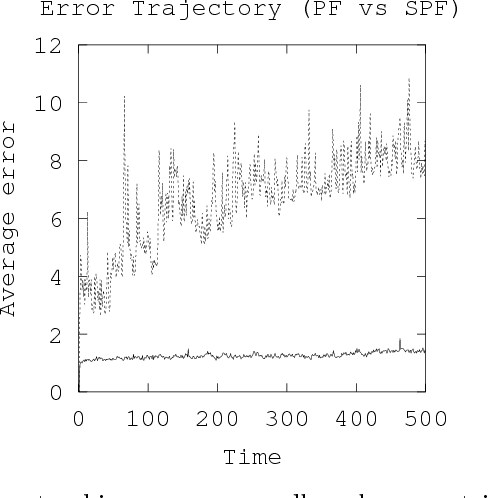
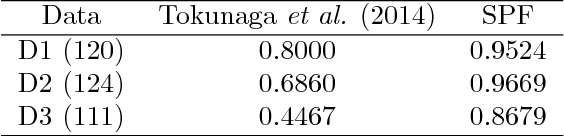
Abstract:Tracking many cells in time-lapse 3D image sequences is an important challenging task of bioimage informatics. Motivated by a study of brain-wide 4D imaging of neural activity in C. elegans, we present a new method of multi-cell tracking. Data types to which the method is applicable are characterized as follows: (i) cells are imaged as globular-like objects, (ii) it is difficult to distinguish cells based only on shape and size, (iii) the number of imaged cells ranges in several hundreds, (iv) moves of nearly-located cells are strongly correlated and (v) cells do not divide. We developed a tracking software suite which we call SPF-CellTracker. Incorporating dependency on cells' moves into prediction model is the key to reduce the tracking errors: cell-switching and coalescence of tracked positions. We model target cells' correlated moves as a Markov random field and we also derive a fast computation algorithm, which we call spatial particle filter. With the live-imaging data of nuclei of C. elegans neurons in which approximately 120 nuclei of neurons are imaged, we demonstrate an advantage of the proposed method over the standard particle filter and a method developed by Tokunaga et al. (2014).
* 14 pages, 6 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge