Osamu Hirose
Dependent landmark drift: robust point set registration with a Gaussian mixture model and a statistical shape model
Jul 26, 2018
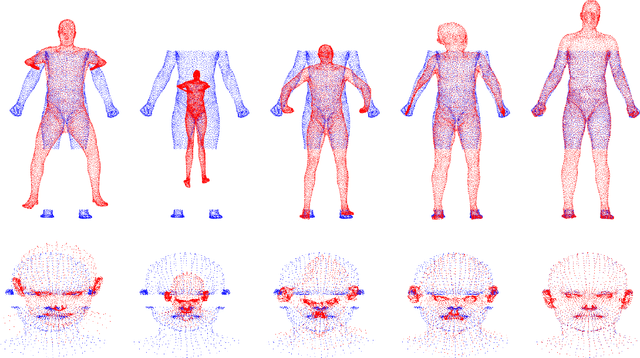
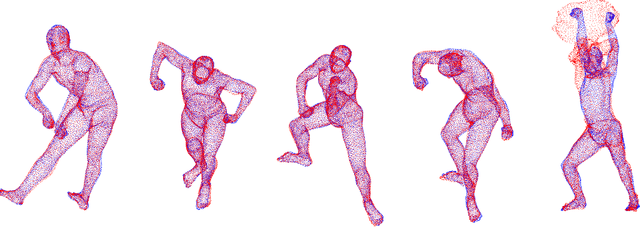
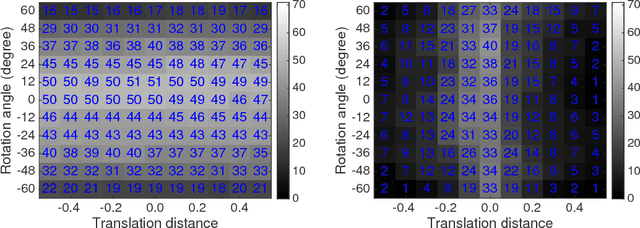
Abstract:The goal of point set registration is to find point-by-point correspondences between point sets, each of which characterizes the shape of an object. Because local preservation of object geometry is assumed, prevalent algorithms in the area can often elegantly solve the problems without using geometric information specific to the objects. This means that registration performance can be further improved by using prior knowledge of object geometry. In this paper, we propose a novel point set registration method using the Gaussian mixture model with prior shape information encoded as a statistical shape model. Our transformation model is defined as a combination of the similar transformation, motion coherence, and the statistical shape model. Therefore, the proposed method works effectively if the target point set includes outliers and missing regions, or if it is rotated. The computational cost can be reduced to linear, and therefore the method is scalable to large point sets. The effectiveness of the method will be verified through comparisons with existing algorithms using datasets concerning human body shapes, hands, and faces.
SPF-CellTracker: Tracking multiple cells with strongly-correlated moves using a spatial particle filter
May 27, 2016
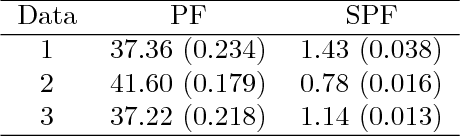
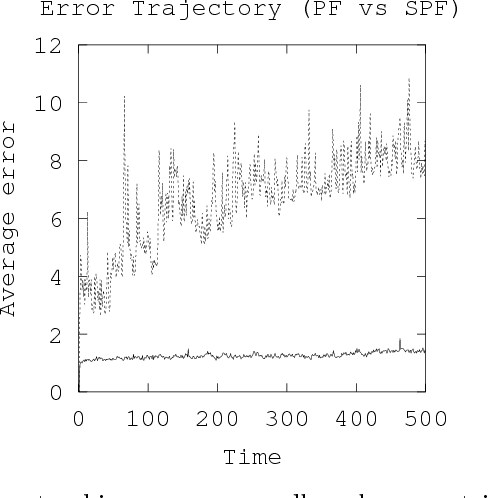
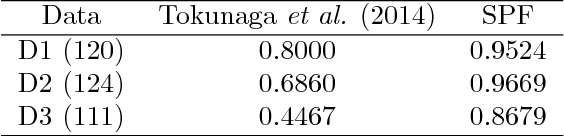
Abstract:Tracking many cells in time-lapse 3D image sequences is an important challenging task of bioimage informatics. Motivated by a study of brain-wide 4D imaging of neural activity in C. elegans, we present a new method of multi-cell tracking. Data types to which the method is applicable are characterized as follows: (i) cells are imaged as globular-like objects, (ii) it is difficult to distinguish cells based only on shape and size, (iii) the number of imaged cells ranges in several hundreds, (iv) moves of nearly-located cells are strongly correlated and (v) cells do not divide. We developed a tracking software suite which we call SPF-CellTracker. Incorporating dependency on cells' moves into prediction model is the key to reduce the tracking errors: cell-switching and coalescence of tracked positions. We model target cells' correlated moves as a Markov random field and we also derive a fast computation algorithm, which we call spatial particle filter. With the live-imaging data of nuclei of C. elegans neurons in which approximately 120 nuclei of neurons are imaged, we demonstrate an advantage of the proposed method over the standard particle filter and a method developed by Tokunaga et al. (2014).
* 14 pages, 6 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge